
Automated plotting for 'modelbased' objects
Source:R/plot.R, R/tinyplot.R, R/visualisation_recipe.R
visualisation_recipe.estimate_predicted.RdMost modelbased objects can be visualized using either the plot()
function, which internally calls the visualisation_recipe() function and
relies on {ggplot2}. There is also a tinyplot() method, which uses the
{tinyplot} package and relies on the core R graphic system. See the
examples below for more information and examples on how to create and
customize plots.
The plotting works by mapping any predictors from the by argument to the
x-axis, colors, alpha (transparency) and facets. Thus, the appearance of the
plot depends on the order of the variables that you specify in the by
argument. For instance, the plots corresponding to
estimate_relation(model, by=c("Species", "Sepal.Length")) and
estimate_relation(model, by=c("Sepal.Length", "Species")) will look
different.
The automated plotting is primarily meant for convenient visual checks, but
for publication-ready figures, we recommend re-creating the figures using the
{ggplot2} package directly.
Usage
# S3 method for class 'estimate_predicted'
plot(x, ...)
# S3 method for class 'estimate_means'
plot(x, ...)
# S3 method for class 'estimate_means'
tinyplot(
x,
type = NULL,
dodge = NULL,
show_data = FALSE,
collapse_group = NULL,
numeric_as_discrete = NULL,
...
)
# S3 method for class 'estimate_predicted'
visualisation_recipe(
x,
show_data = FALSE,
show_residuals = FALSE,
collapse_group = NULL,
point = NULL,
line = NULL,
pointrange = NULL,
ribbon = NULL,
facet = NULL,
grid = NULL,
join_dots = NULL,
numeric_as_discrete = NULL,
...
)
# S3 method for class 'estimate_slopes'
visualisation_recipe(
x,
line = NULL,
pointrange = NULL,
ribbon = NULL,
facet = NULL,
grid = NULL,
...
)
# S3 method for class 'estimate_grouplevel'
visualisation_recipe(
x,
line = NULL,
pointrange = NULL,
ribbon = NULL,
facet = NULL,
grid = NULL,
...
)Arguments
- x
A modelbased object.
- ...
Arguments passed from
plot()tovisualisation_recipe(), or totinyplot()if you use that method.- type
The type of
tinyplotvisualization. It is recommended that users leave asNULL(the default), in which case the plot type will be determined automatically by the underlyingmodelbasedobject.- dodge
Dodge value for grouped plots. If
NULL(the default), then the dodging behavior is determined by the number of groups andgetOption("modelbased_tinyplot_dodge").- show_data
Logical, if
TRUE, display the "raw" data as a background to the model-based estimation. For mixed models, you can additionally use thecollapse_groupargument to "collapse" data points by random effects grouping factors. Argumentshow_datawill be ignored for plotting objects returned byestimate_slopes()orestimate_grouplevel().- collapse_group
This argument only takes effect when either
show_dataorshow_residualsisTRUE. For mixed effects models, name of the grouping variable of random effects. Ifcollapse_group = TRUE, data points "collapsed" by the first random effect groups are added to the plot. Else, ifcollapse_groupis a name of a group factor, data is collapsed by that specific random effect. Seecollapse_by_group()for further details.- numeric_as_discrete
Maximum number of unique values in a numeric predictor to treat that predictor as discrete. Defaults to
8. Numeric predictors are usually mapped to a continuous color scale, unless they have only few unique values. In the latter case, numeric predictors are assumed to represent "categories", e.g. when only the mean value and +/- 1 standard deviation around the mean are chosen as representative values for that predictor. UseFALSEto always use continuous color scales for numeric predictors. It is possible to set a global default value usingoptions(), e.g.options(modelbased_numeric_as_discrete = 10).- show_residuals
Logical, if
TRUE, display residuals of the model as a background to the model-based estimation. Residuals will be computed for the predictors in the data grid, usingresidualize_over_grid(). For mixed models, you can additionally use thecollapse_groupargument to "collapse" data points from residuals by random effects grouping factors.- point, line, pointrange, ribbon, facet, grid
Additional aesthetics and parameters for the geoms (see customization example).
- join_dots
Logical, if
TRUE(default) and for categorical focal terms inby, dots (estimates) are connected by lines, i.e. plots will be a combination of dots with error bars and connecting lines. IfFALSE, only dots and error bars are shown. It is possible to set a global default value usingoptions(), e.g.options(modelbased_join_dots = FALSE).
Value
An object of class visualisation_recipe that describes the layers
used to create a plot based on {ggplot2}. The related plot() method is in
the {see} package.
Details
There are two options to remove the confidence bands or errors bars
from the plot. To remove error bars, simply set the pointrange geom to
point, e.g. plot(..., pointrange = list(geom = "point")). To remove the
confidence bands from line geoms, use ribbon = "none".
Global Options to Customize Plots
Some arguments for plot() can get global defaults using options():
modelbased_join_dots:options(modelbased_join_dots = <logical>)will set a default value for thejoin_dots.modelbased_numeric_as_discrete:options(modelbased_numeric_as_discrete = <number>)will set a default value for themodelbased_numeric_as_discreteargument. Can also beFALSE.modelbased_ribbon_alpha:options(modelbased_ribbon_alpha = <number>)will set a default value for thealphaargument of theribbongeom. Should be a number between0and1.modelbased_tinyplot_dodge:options(modelbased_tinyplot_dodge = <number>)will set a default value for thedodgeargument (spacing between geoms) when usingtinyplot::plt(). Should be a number between0and1.
Examples
# ==============================================
# tinyplot
# ==============================================
# \donttest{
library(tinyplot)
data(efc, package = "modelbased")
efc <- datawizard::to_factor(efc, c("e16sex", "c172code", "e42dep"))
m <- lm(neg_c_7 ~ e16sex + c172code + barthtot, data = efc)
em <- estimate_means(m, "c172code")
plt(em)
 # pass additional tinyplot arguments for customization, e.g.
plt(em, theme = "classic")
# pass additional tinyplot arguments for customization, e.g.
plt(em, theme = "classic")
 plt(em, theme = "classic", flip = TRUE)
plt(em, theme = "classic", flip = TRUE)
 # etc.
# Aside: use tinyplot::tinytheme() to set a persistent theme
tinytheme("classic")
# continuous variable example
em <- estimate_means(m, "barthtot")
plt(em)
# etc.
# Aside: use tinyplot::tinytheme() to set a persistent theme
tinytheme("classic")
# continuous variable example
em <- estimate_means(m, "barthtot")
plt(em)
 # grouped example
m <- lm(neg_c_7 ~ e16sex * c172code + e42dep, data = efc)
em <- estimate_means(m, c("e16sex", "c172code"))
plt(em)
# use plt_add (alias tinyplot_add) to add layers
plt_add(type = "l", lty = 2)
# grouped example
m <- lm(neg_c_7 ~ e16sex * c172code + e42dep, data = efc)
em <- estimate_means(m, c("e16sex", "c172code"))
plt(em)
# use plt_add (alias tinyplot_add) to add layers
plt_add(type = "l", lty = 2)
 # Reset to default theme
tinytheme()
# }
library(ggplot2)
library(see)
# ==============================================
# estimate_relation, estimate_expectation, ...
# ==============================================
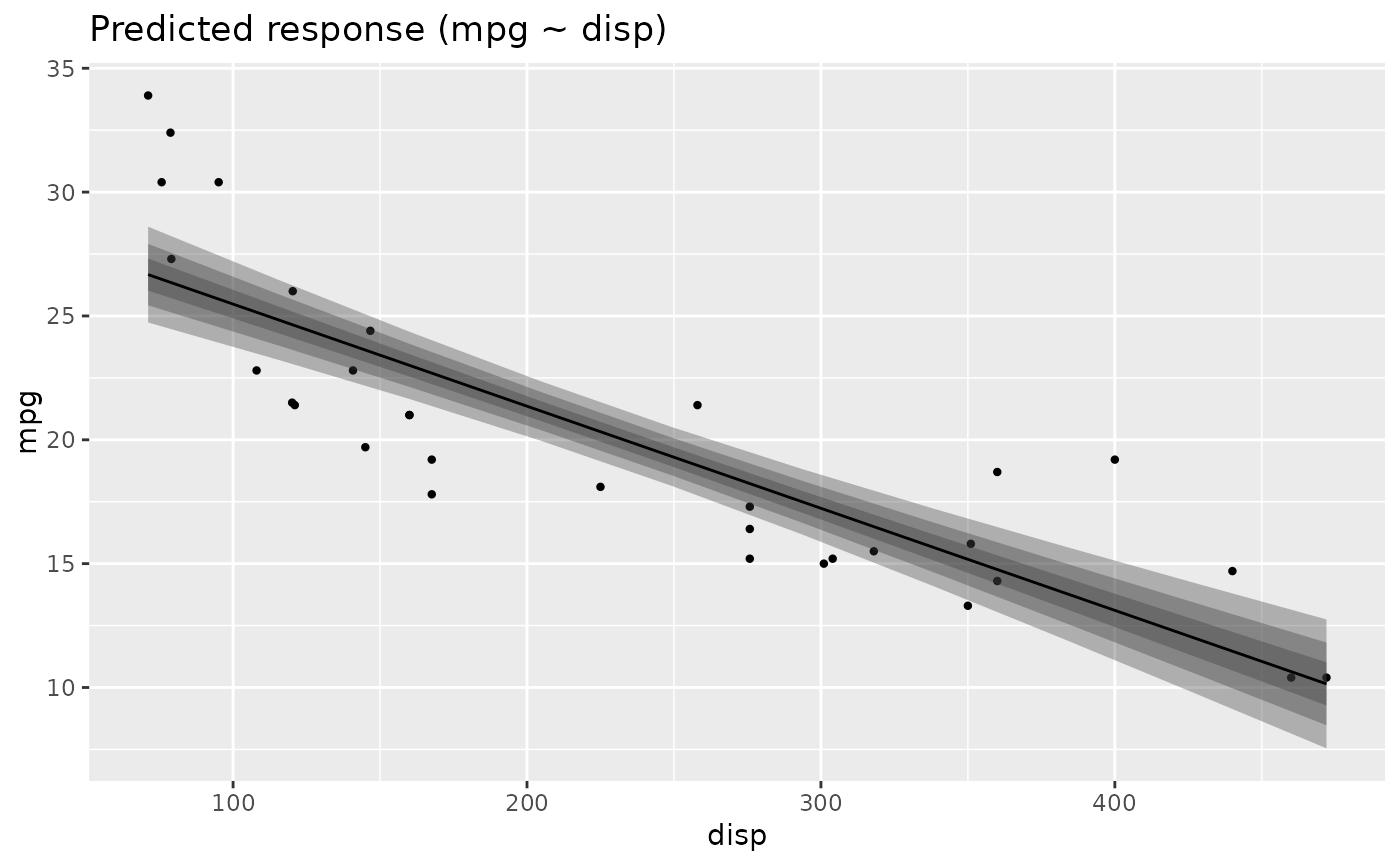
# Simple Model ---------------
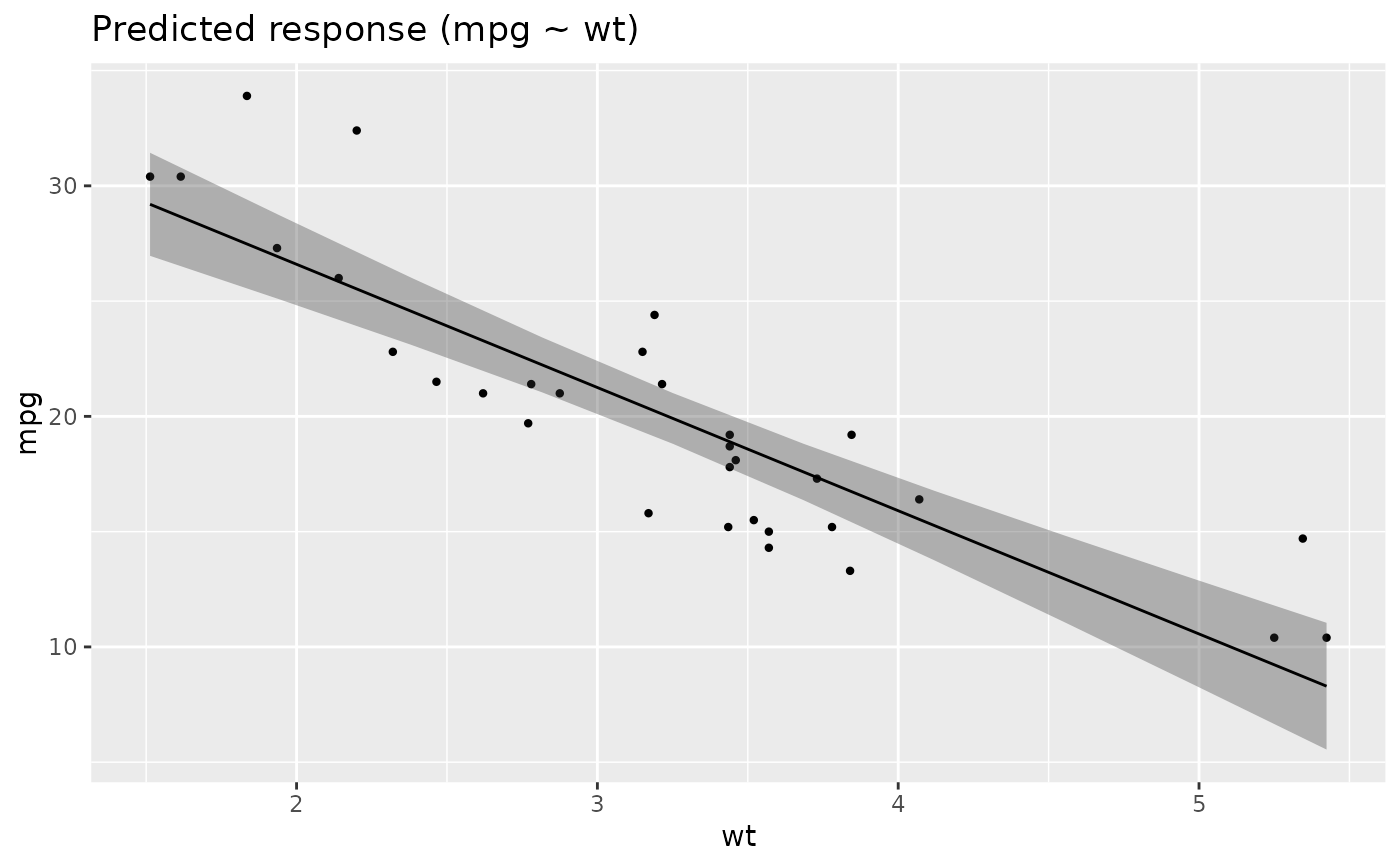
x <- estimate_relation(lm(mpg ~ wt, data = mtcars))
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: ribbon
#> data = [10 x 6]
#> aes_string(
#> y = 'Predicted'
#> x = 'wt'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#> alpha = 0.25
#>
#> Layer 2
#> --------
#> Geom type: line
#> data = [10 x 6]
#> aes_string(
#> y = 'Predicted'
#> x = 'wt'
#> group = '.group'
#> )
#>
#> Layer 3
#> --------
#> Geom type: labs
#> y = 'Predicted value of mpg'
#>
plot(layers)
# Reset to default theme
tinytheme()
# }
library(ggplot2)
library(see)
# ==============================================
# estimate_relation, estimate_expectation, ...
# ==============================================
# Simple Model ---------------
x <- estimate_relation(lm(mpg ~ wt, data = mtcars))
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: ribbon
#> data = [10 x 6]
#> aes_string(
#> y = 'Predicted'
#> x = 'wt'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#> alpha = 0.25
#>
#> Layer 2
#> --------
#> Geom type: line
#> data = [10 x 6]
#> aes_string(
#> y = 'Predicted'
#> x = 'wt'
#> group = '.group'
#> )
#>
#> Layer 3
#> --------
#> Geom type: labs
#> y = 'Predicted value of mpg'
#>
plot(layers)
 # visualization_recipe() is called implicitly when you call plot()
plot(estimate_relation(lm(mpg ~ qsec, data = mtcars)))
# visualization_recipe() is called implicitly when you call plot()
plot(estimate_relation(lm(mpg ~ qsec, data = mtcars)))
 # \dontrun{
# It can be used in a pipe workflow
lm(mpg ~ qsec, data = mtcars) |>
estimate_relation(ci = c(0.5, 0.8, 0.9)) |>
plot()
# \dontrun{
# It can be used in a pipe workflow
lm(mpg ~ qsec, data = mtcars) |>
estimate_relation(ci = c(0.5, 0.8, 0.9)) |>
plot()
 # Customize aesthetics ----------
plot(x,
point = list(color = "red", alpha = 0.6, size = 3),
line = list(color = "blue", size = 3),
ribbon = list(fill = "green", alpha = 0.7)
) +
theme_minimal() +
labs(title = "Relationship between MPG and WT")
# Customize aesthetics ----------
plot(x,
point = list(color = "red", alpha = 0.6, size = 3),
line = list(color = "blue", size = 3),
ribbon = list(fill = "green", alpha = 0.7)
) +
theme_minimal() +
labs(title = "Relationship between MPG and WT")
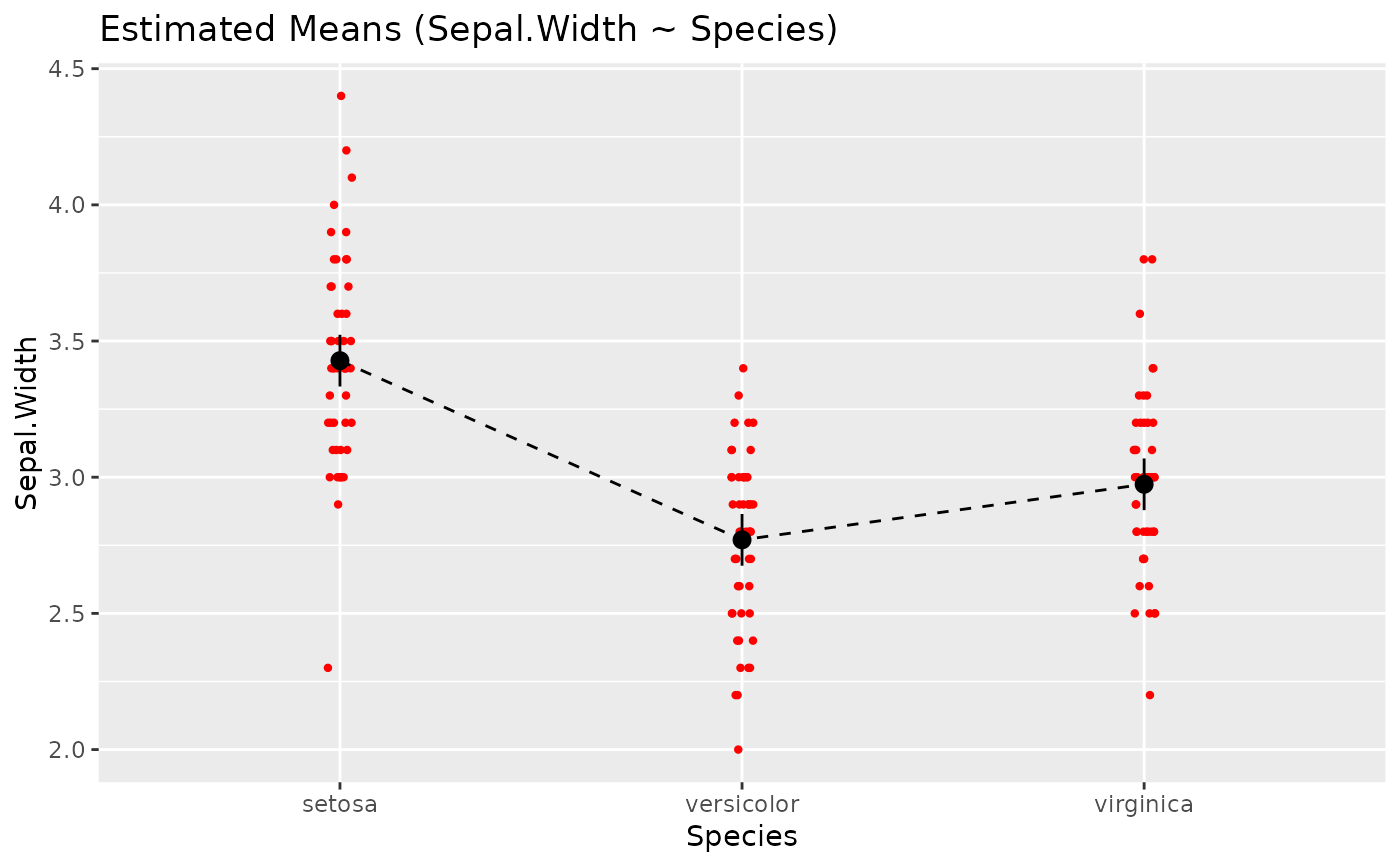
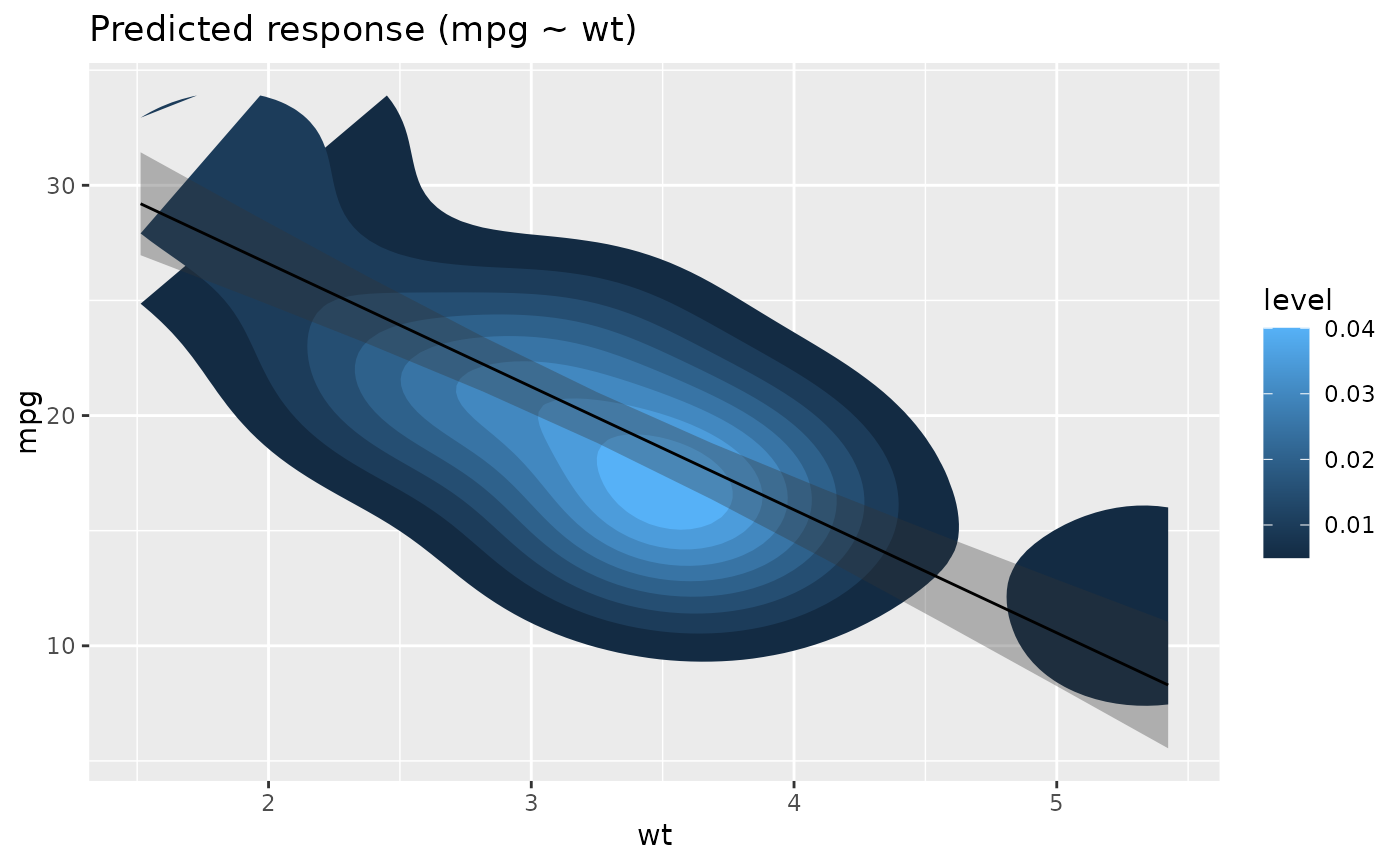
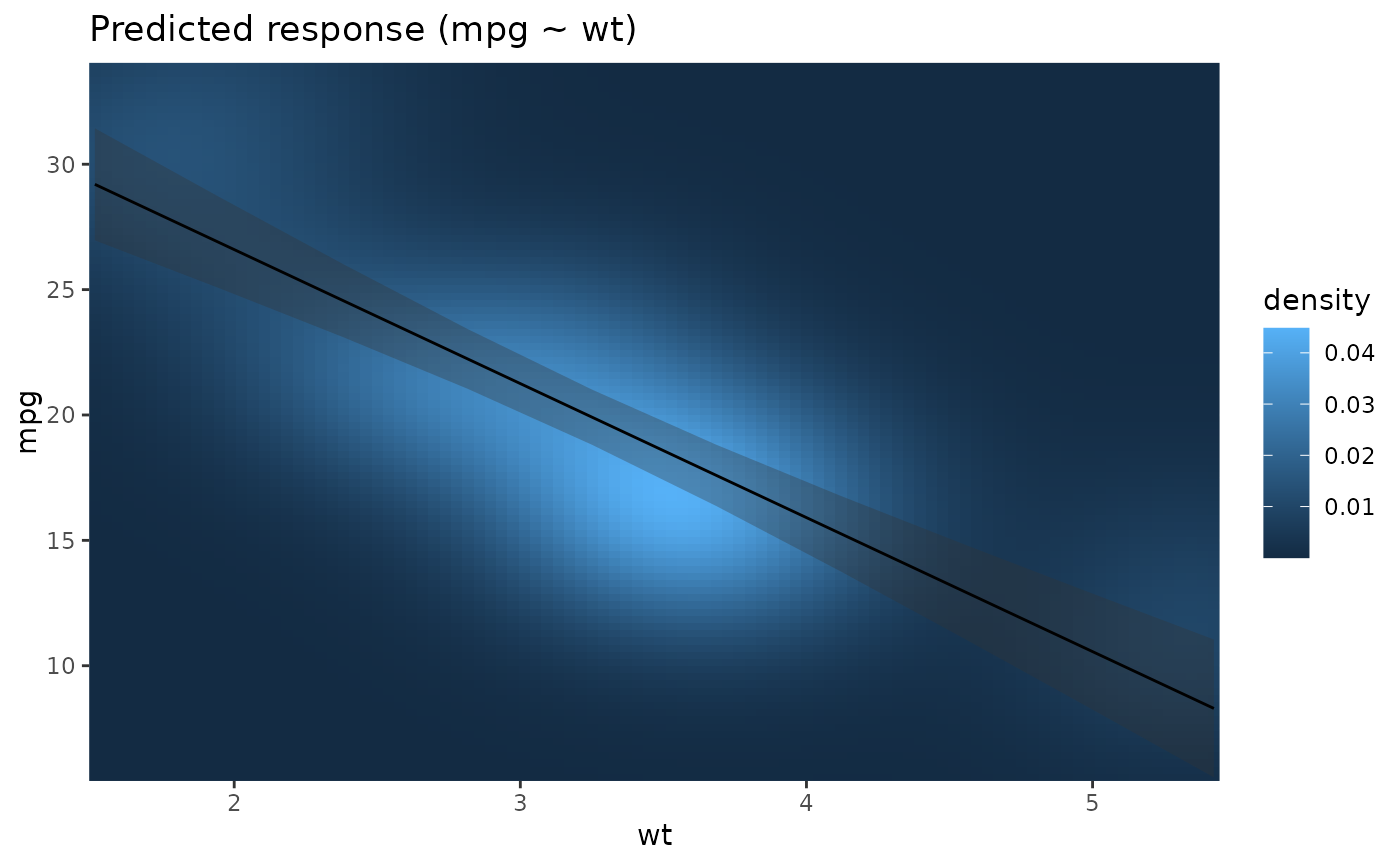
 # Customize raw data -------------
plot(x, point = list(geom = "density_2d_filled"), line = list(color = "white")) +
scale_x_continuous(expand = c(0, 0)) +
scale_y_continuous(expand = c(0, 0)) +
theme(legend.position = "none")
# Customize raw data -------------
plot(x, point = list(geom = "density_2d_filled"), line = list(color = "white")) +
scale_x_continuous(expand = c(0, 0)) +
scale_y_continuous(expand = c(0, 0)) +
theme(legend.position = "none")
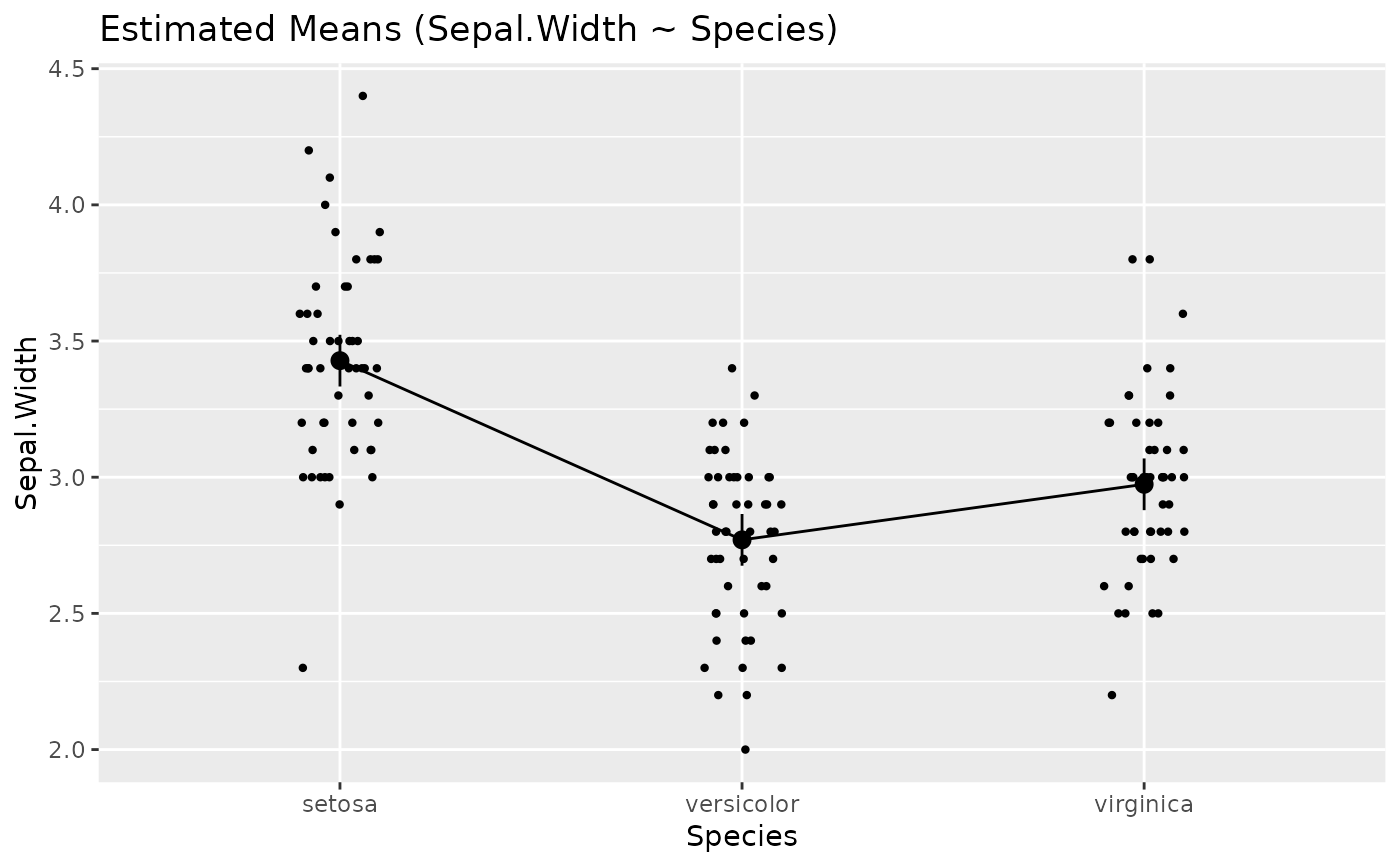
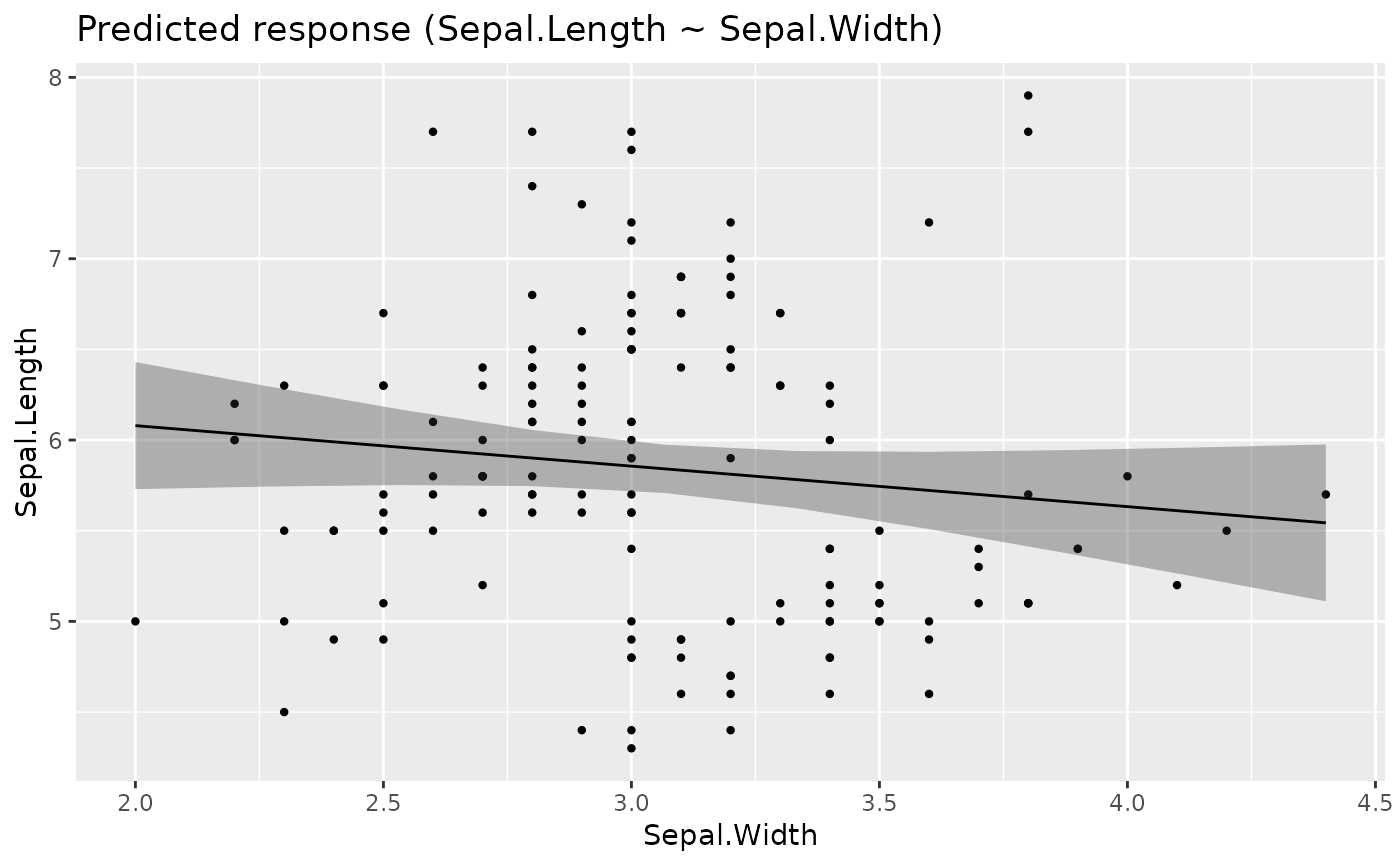
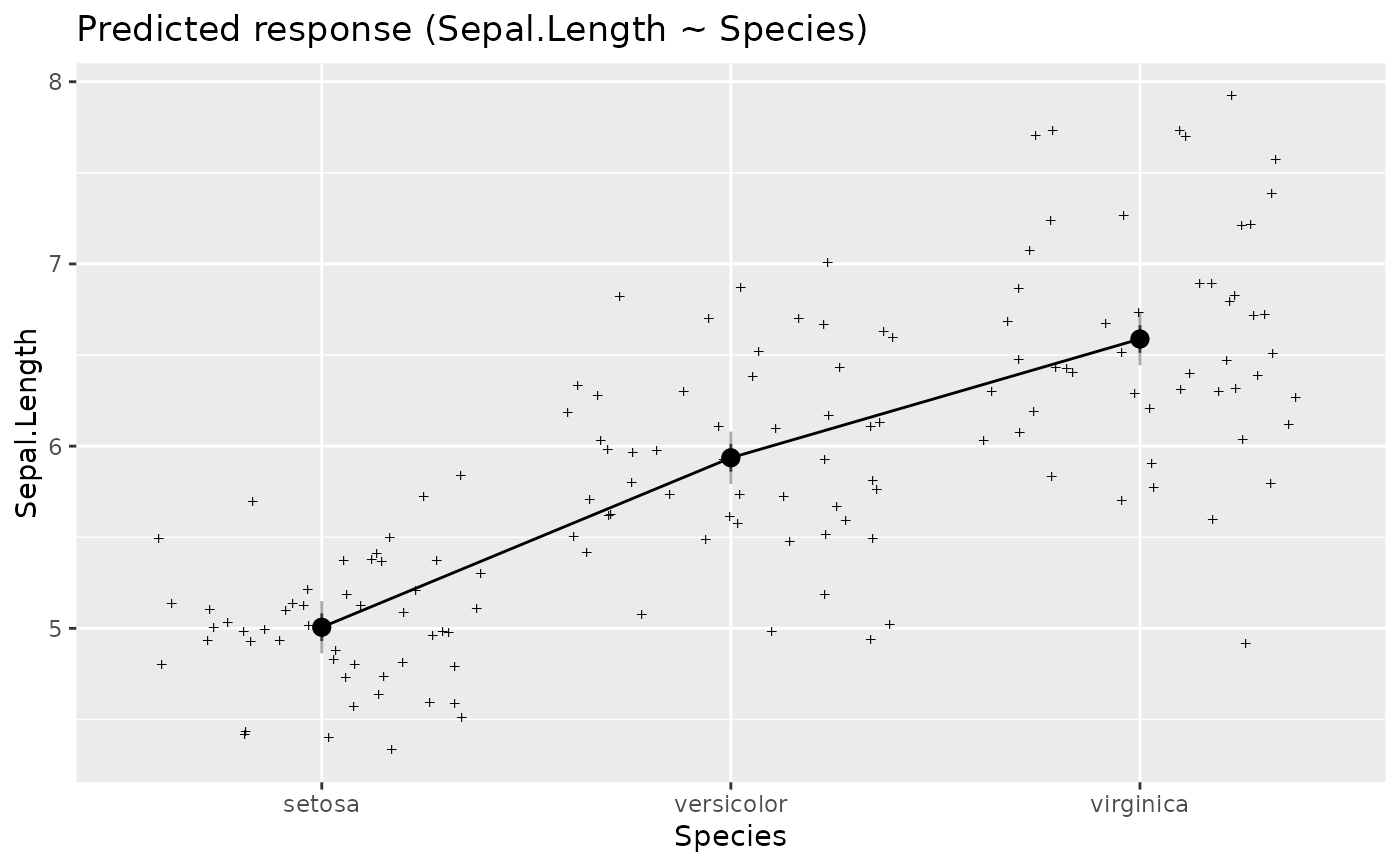 # Single predictors examples -----------
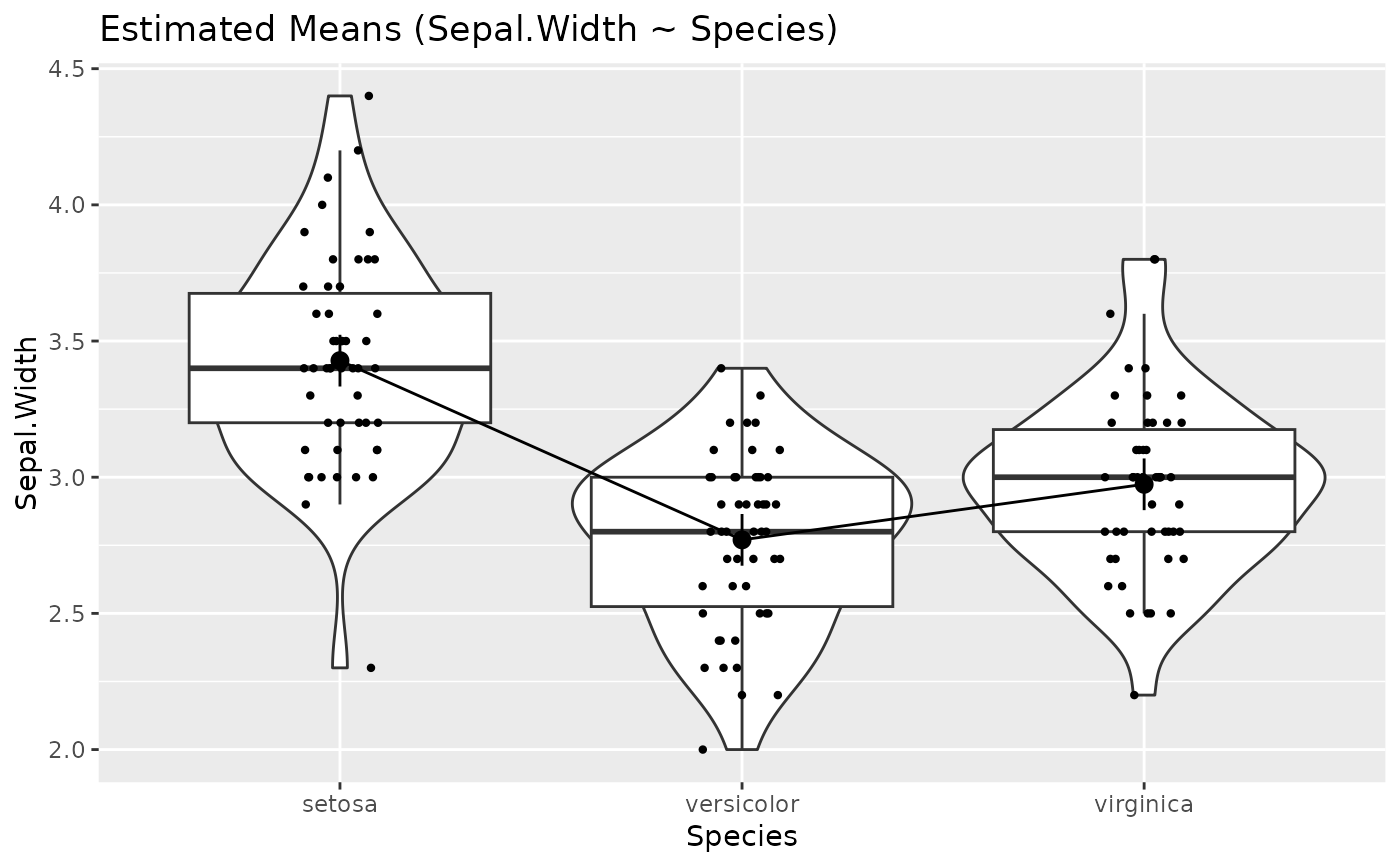
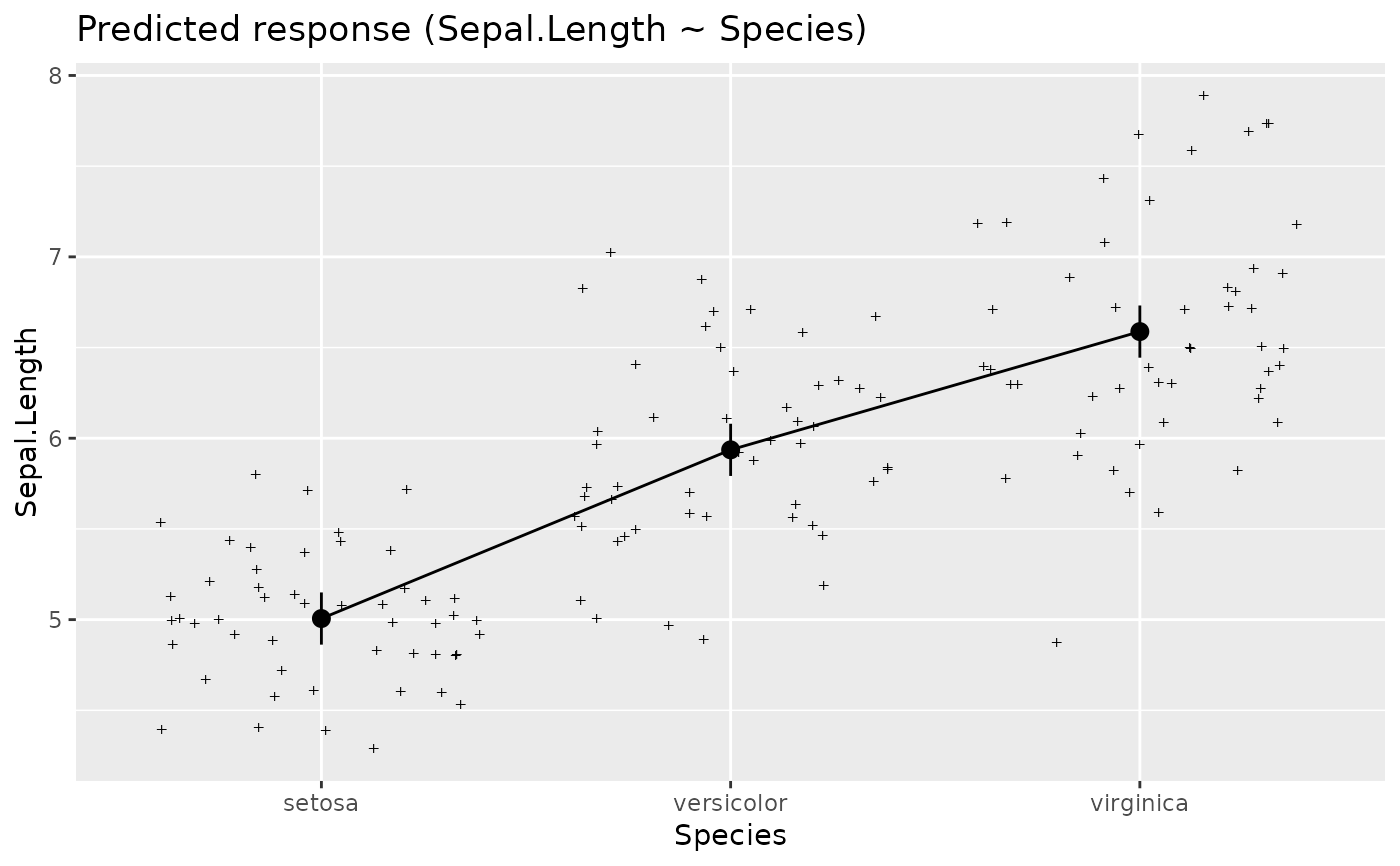
plot(estimate_relation(lm(Sepal.Length ~ Species, data = iris)))
# Single predictors examples -----------
plot(estimate_relation(lm(Sepal.Length ~ Species, data = iris)))
 # 2-ways interaction ------------
# Numeric * numeric
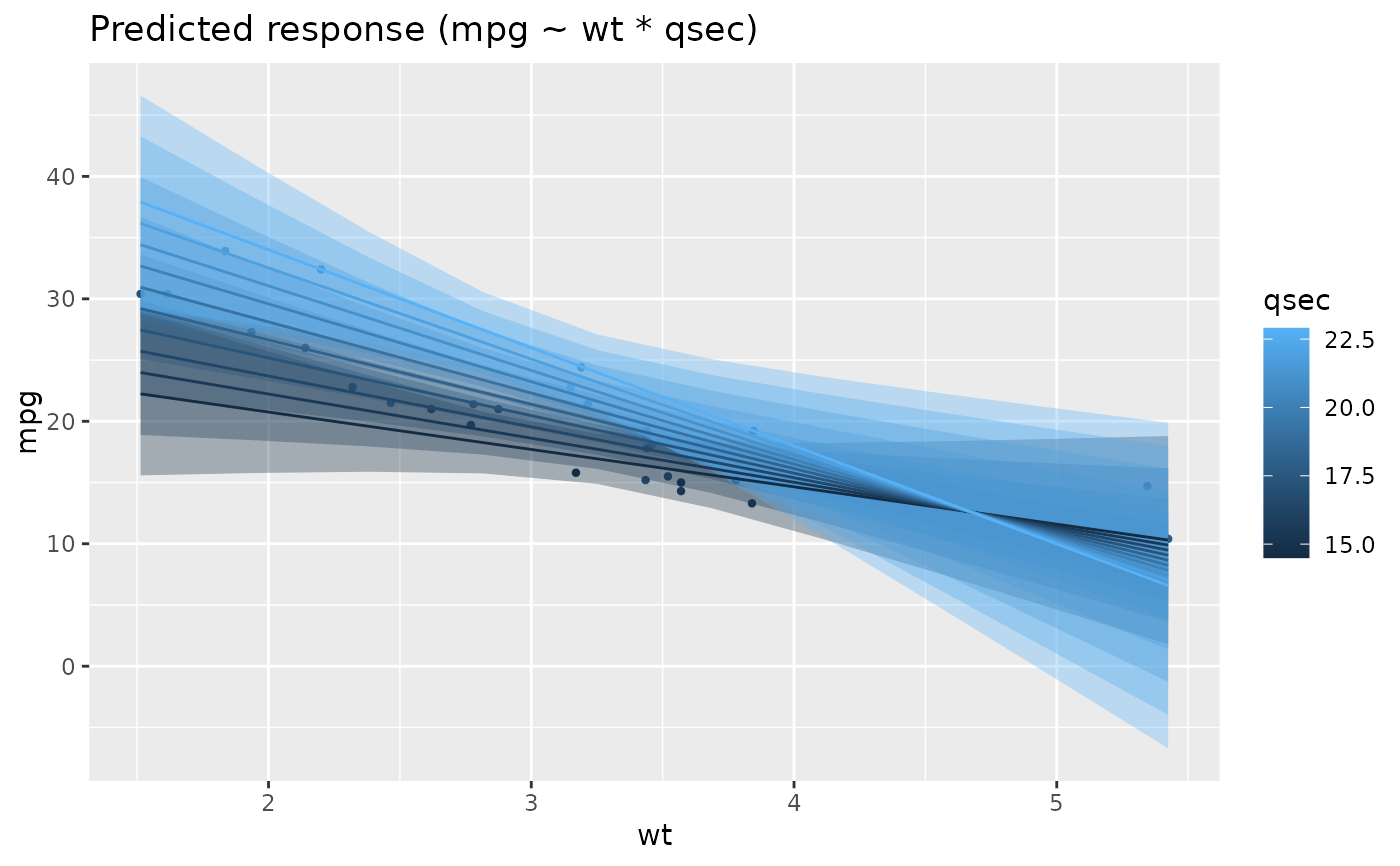
x <- estimate_relation(lm(mpg ~ wt * qsec, data = mtcars))
plot(x)
# 2-ways interaction ------------
# Numeric * numeric
x <- estimate_relation(lm(mpg ~ wt * qsec, data = mtcars))
plot(x)
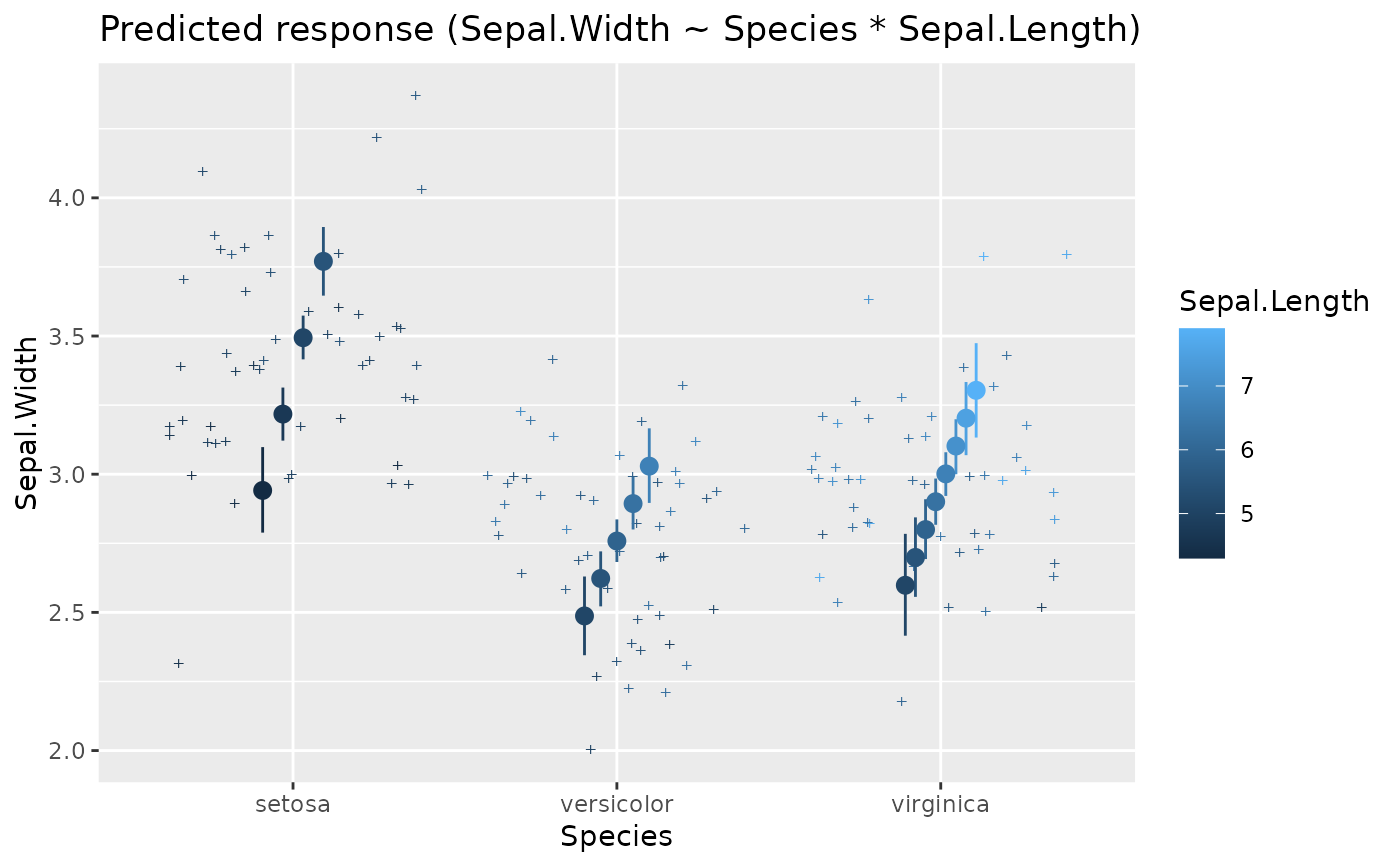
 # Numeric * factor
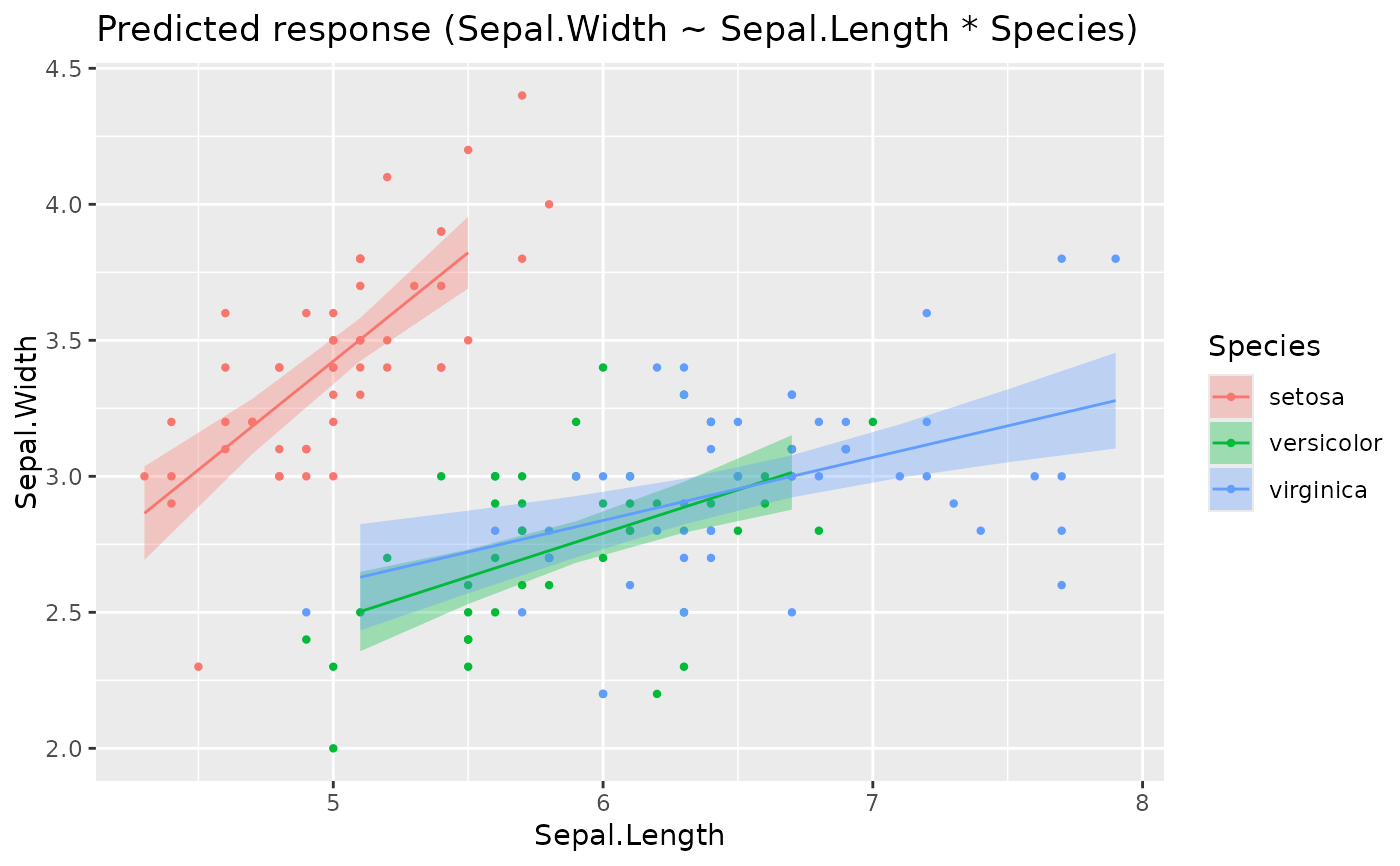
x <- estimate_relation(lm(Sepal.Width ~ Sepal.Length * Species, data = iris))
plot(x)
# Numeric * factor
x <- estimate_relation(lm(Sepal.Width ~ Sepal.Length * Species, data = iris))
plot(x)
 # ==============================================
# estimate_means
# ==============================================
# Simple Model ---------------
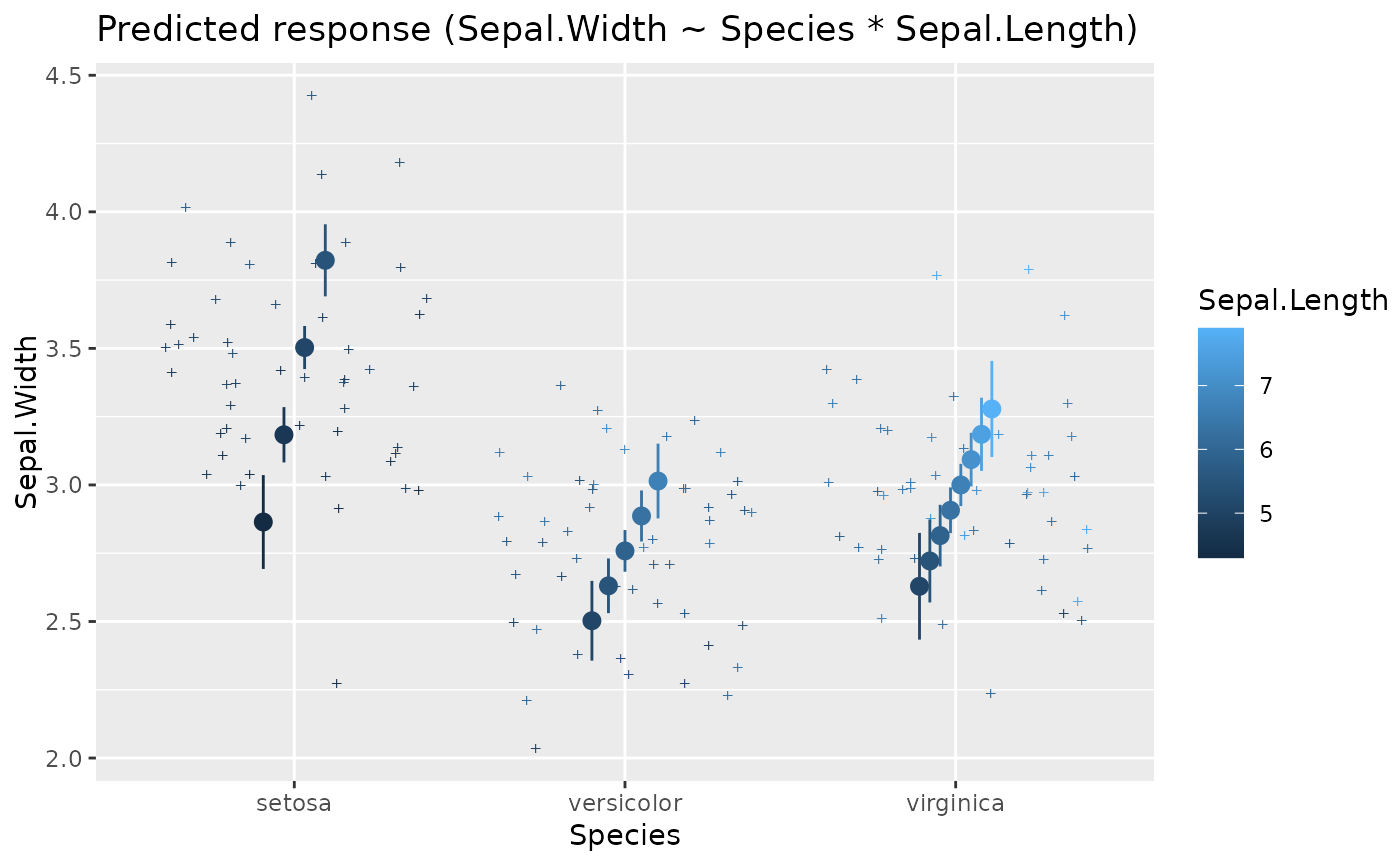
x <- estimate_means(lm(Sepal.Width ~ Species, data = iris), by = "Species")
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: line
#> data = [3 x 8]
#> aes_string(
#> y = 'Mean'
#> x = 'Species'
#> group = '.group'
#> )
#>
#> Layer 2
#> --------
#> Geom type: pointrange
#> data = [3 x 8]
#> aes_string(
#> y = 'Mean'
#> x = 'Species'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#>
#> Layer 3
#> --------
#> Geom type: labs
#> y = 'Mean of Sepal.Width'
#>
plot(layers)
# ==============================================
# estimate_means
# ==============================================
# Simple Model ---------------
x <- estimate_means(lm(Sepal.Width ~ Species, data = iris), by = "Species")
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: line
#> data = [3 x 8]
#> aes_string(
#> y = 'Mean'
#> x = 'Species'
#> group = '.group'
#> )
#>
#> Layer 2
#> --------
#> Geom type: pointrange
#> data = [3 x 8]
#> aes_string(
#> y = 'Mean'
#> x = 'Species'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#>
#> Layer 3
#> --------
#> Geom type: labs
#> y = 'Mean of Sepal.Width'
#>
plot(layers)
 # Customize aesthetics
layers <- visualisation_recipe(x,
point = list(width = 0.03, color = "red"),
pointrange = list(size = 2, linewidth = 2),
line = list(linetype = "dashed", color = "blue")
)
plot(layers)
# Customize aesthetics
layers <- visualisation_recipe(x,
point = list(width = 0.03, color = "red"),
pointrange = list(size = 2, linewidth = 2),
line = list(linetype = "dashed", color = "blue")
)
plot(layers)
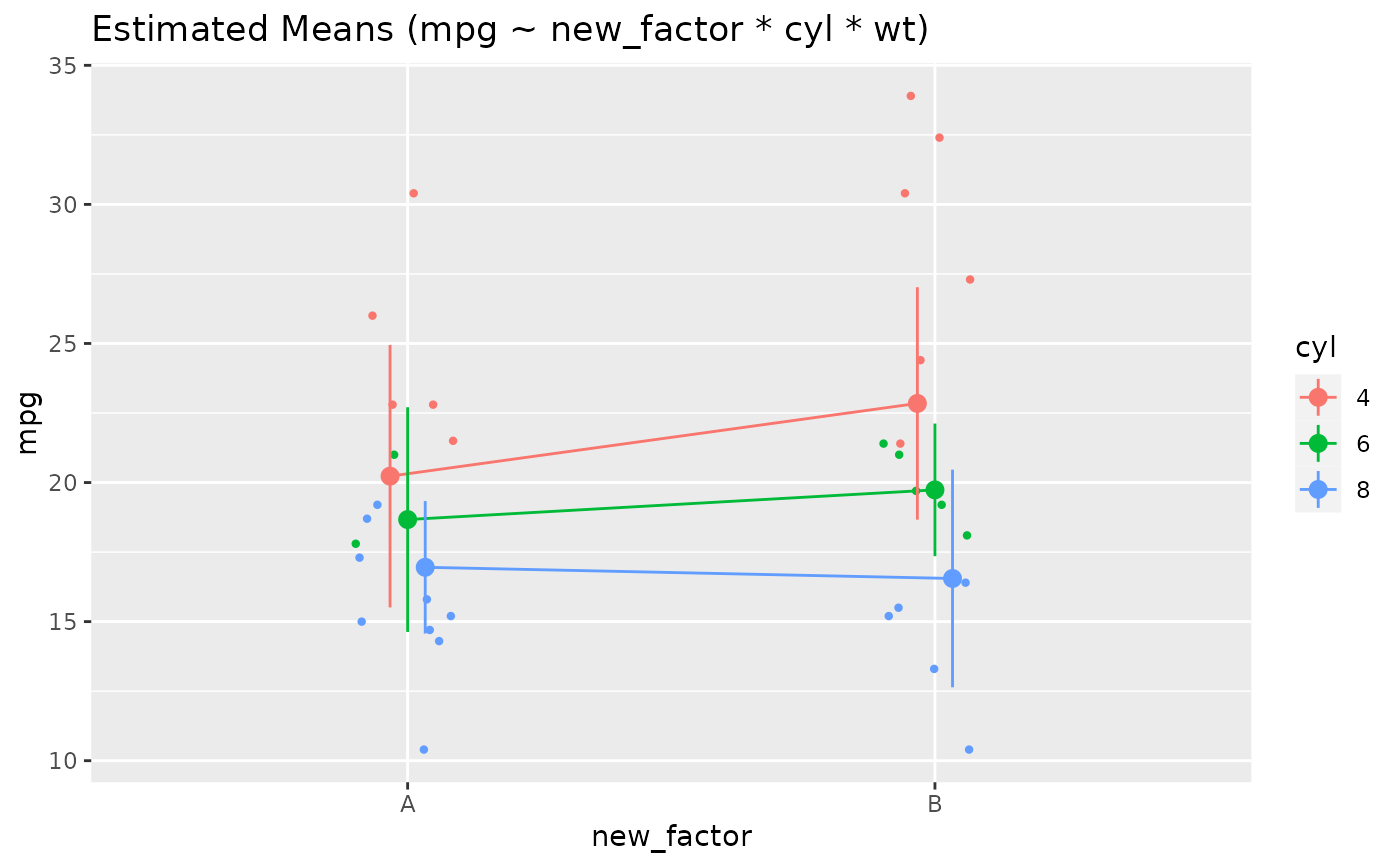
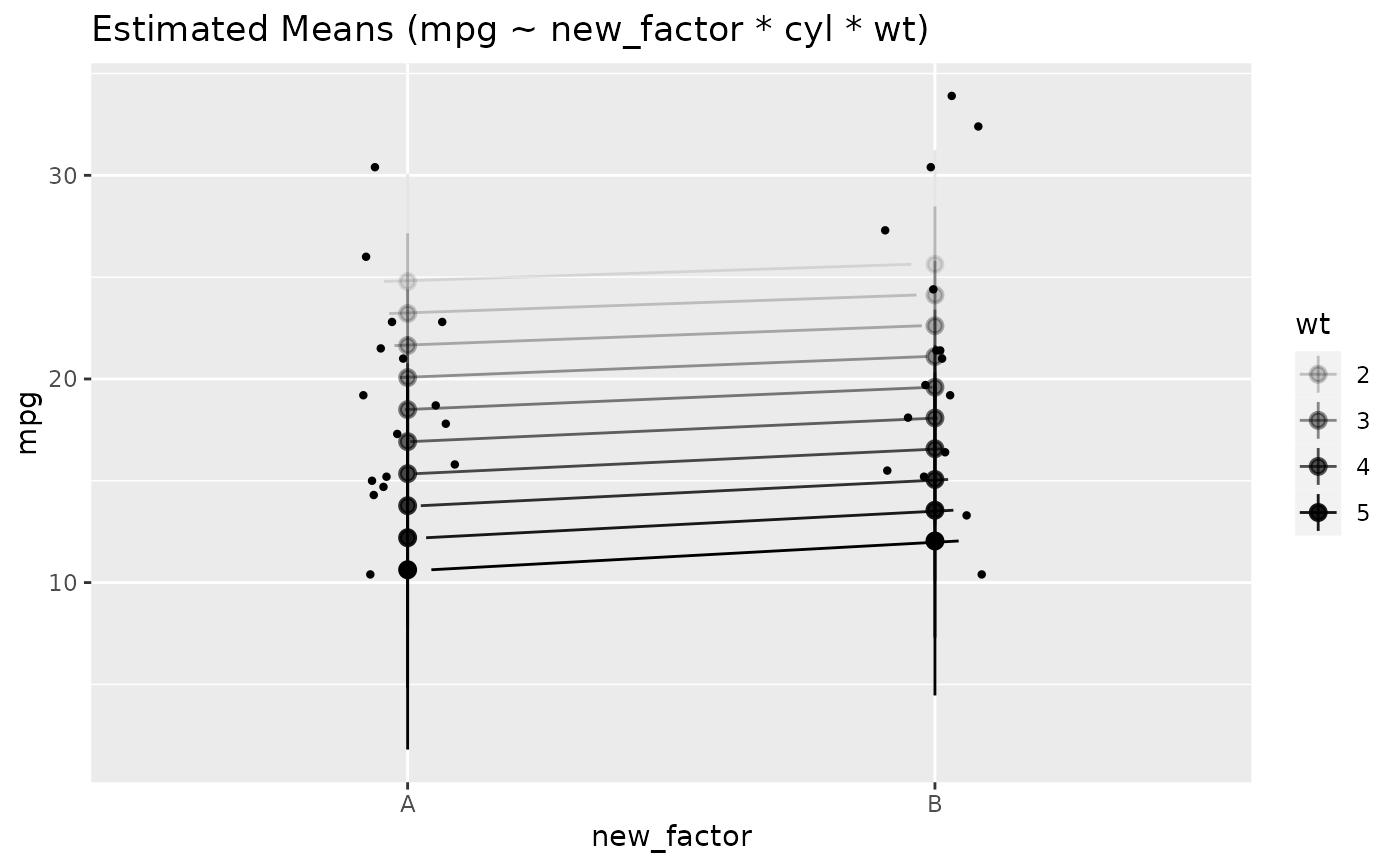
 # Two levels ---------------
data <- mtcars
data$cyl <- as.factor(data$cyl)
model <- lm(mpg ~ cyl * wt, data = data)
x <- estimate_means(model, by = c("cyl", "wt"))
plot(x)
# Two levels ---------------
data <- mtcars
data$cyl <- as.factor(data$cyl)
model <- lm(mpg ~ cyl * wt, data = data)
x <- estimate_means(model, by = c("cyl", "wt"))
plot(x)
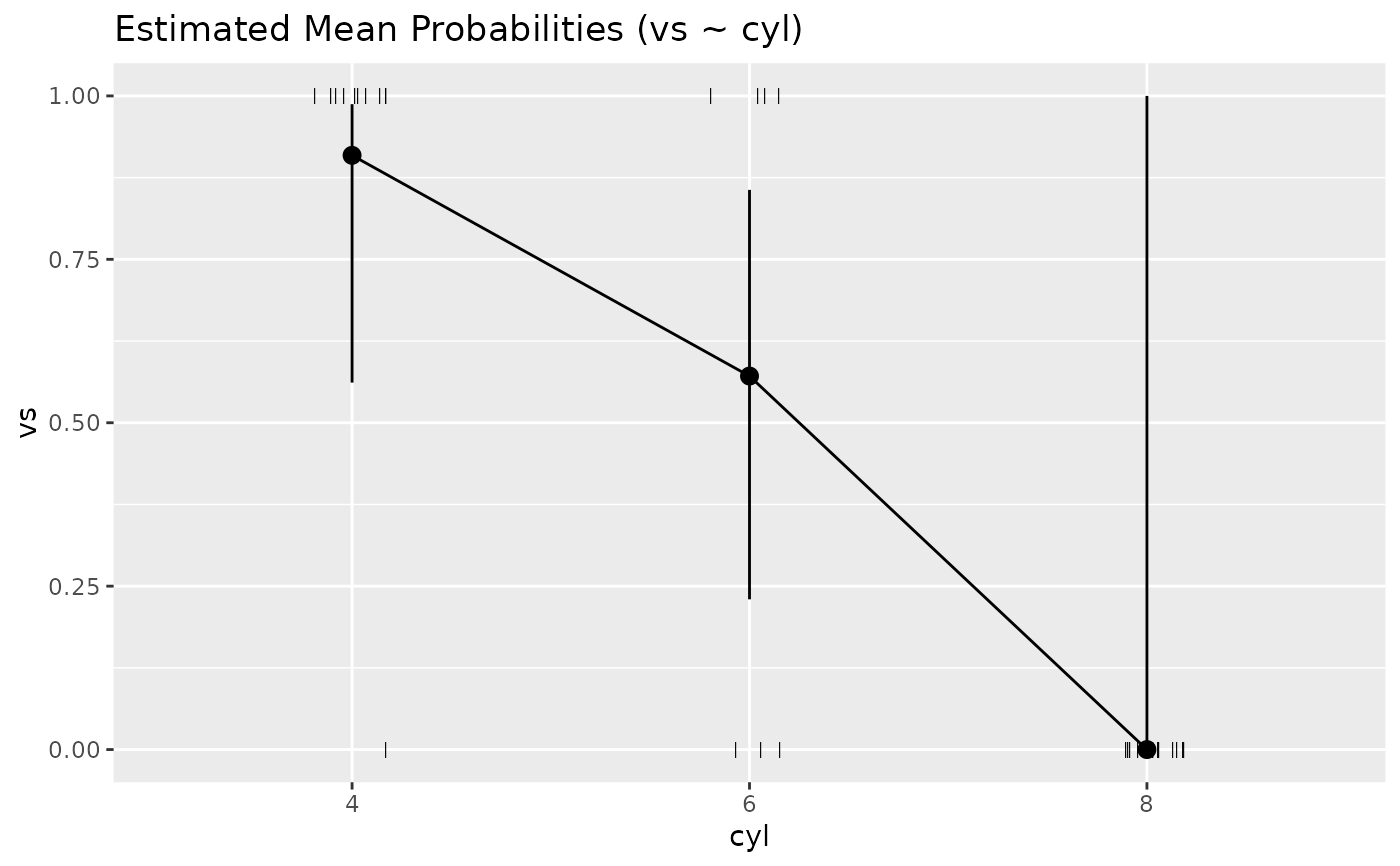
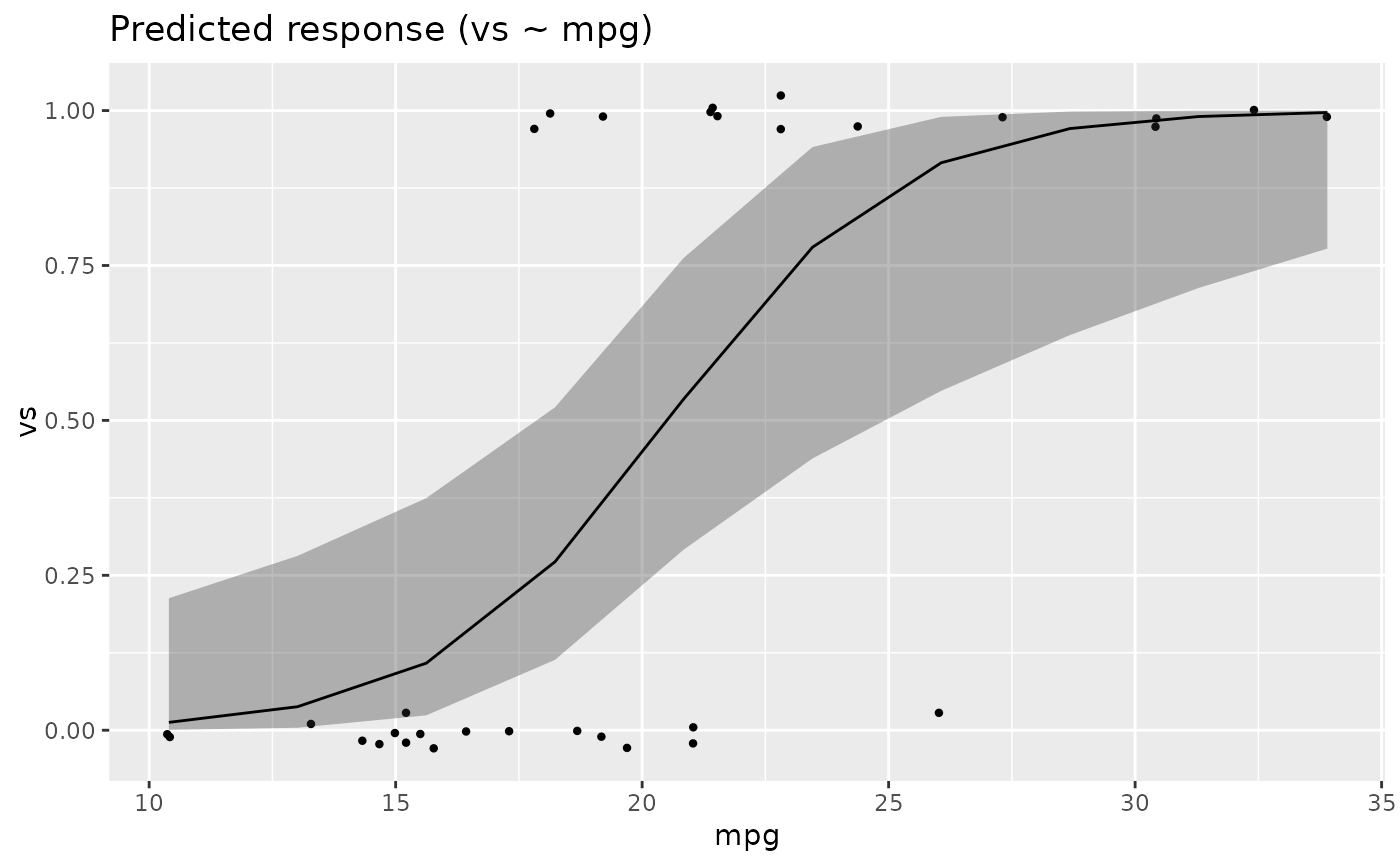
 # GLMs ---------------------
data <- data.frame(vs = mtcars$vs, cyl = as.factor(mtcars$cyl))
x <- estimate_means(glm(vs ~ cyl, data = data, family = "binomial"), by = c("cyl"))
plot(x)
# GLMs ---------------------
data <- data.frame(vs = mtcars$vs, cyl = as.factor(mtcars$cyl))
x <- estimate_means(glm(vs ~ cyl, data = data, family = "binomial"), by = c("cyl"))
plot(x)
 # ==============================================
# Adding original data to the plot
# ==============================================
data(efc, package = "modelbased")
efc$e15relat <- as.factor(efc$e15relat)
efc$c161sex <- as.factor(efc$c161sex)
levels(efc$c161sex) <- c("male", "female")
model <- lme4::lmer(neg_c_7 ~ c161sex + (1 | e15relat), data = efc)
me <- estimate_means(model, "c161sex")
plot(me, show_data = TRUE)
# ==============================================
# Adding original data to the plot
# ==============================================
data(efc, package = "modelbased")
efc$e15relat <- as.factor(efc$e15relat)
efc$c161sex <- as.factor(efc$c161sex)
levels(efc$c161sex) <- c("male", "female")
model <- lme4::lmer(neg_c_7 ~ c161sex + (1 | e15relat), data = efc)
me <- estimate_means(model, "c161sex")
plot(me, show_data = TRUE)
 # data points: collapse by / average over random effects groups -------
plot(me, show_data = TRUE, collapse_group = "e15relat")
# data points: collapse by / average over random effects groups -------
plot(me, show_data = TRUE, collapse_group = "e15relat")
 # }
# ==============================================
# estimate_slopes
# ==============================================
model <- lm(Sepal.Width ~ Species * Petal.Length, data = iris)
x <- estimate_slopes(model, trend = "Petal.Length", by = "Species")
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: hline
#> yintercept = 0
#> alpha = 0.5
#> linetype = 'dashed'
#>
#> Layer 2
#> --------
#> Geom type: line
#> data = [3 x 9]
#> aes_string(
#> y = 'Slope'
#> x = 'Species'
#> group = '.group'
#> )
#>
#> Layer 3
#> --------
#> Geom type: pointrange
#> data = [3 x 9]
#> aes_string(
#> y = 'Slope'
#> x = 'Species'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#>
#> Layer 4
#> --------
#> Geom type: labs
#> y = 'Slope of Petal.Length'
#>
plot(layers)
# }
# ==============================================
# estimate_slopes
# ==============================================
model <- lm(Sepal.Width ~ Species * Petal.Length, data = iris)
x <- estimate_slopes(model, trend = "Petal.Length", by = "Species")
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: hline
#> yintercept = 0
#> alpha = 0.5
#> linetype = 'dashed'
#>
#> Layer 2
#> --------
#> Geom type: line
#> data = [3 x 9]
#> aes_string(
#> y = 'Slope'
#> x = 'Species'
#> group = '.group'
#> )
#>
#> Layer 3
#> --------
#> Geom type: pointrange
#> data = [3 x 9]
#> aes_string(
#> y = 'Slope'
#> x = 'Species'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#>
#> Layer 4
#> --------
#> Geom type: labs
#> y = 'Slope of Petal.Length'
#>
plot(layers)
 # \dontrun{
# Customize aesthetics and add horizontal line and theme
layers <- visualisation_recipe(x, pointrange = list(size = 2, linewidth = 2))
plot(layers) +
geom_hline(yintercept = 0, linetype = "dashed", color = "red") +
theme_minimal() +
labs(y = "Effect of Petal.Length", title = "Marginal Effects")
# \dontrun{
# Customize aesthetics and add horizontal line and theme
layers <- visualisation_recipe(x, pointrange = list(size = 2, linewidth = 2))
plot(layers) +
geom_hline(yintercept = 0, linetype = "dashed", color = "red") +
theme_minimal() +
labs(y = "Effect of Petal.Length", title = "Marginal Effects")
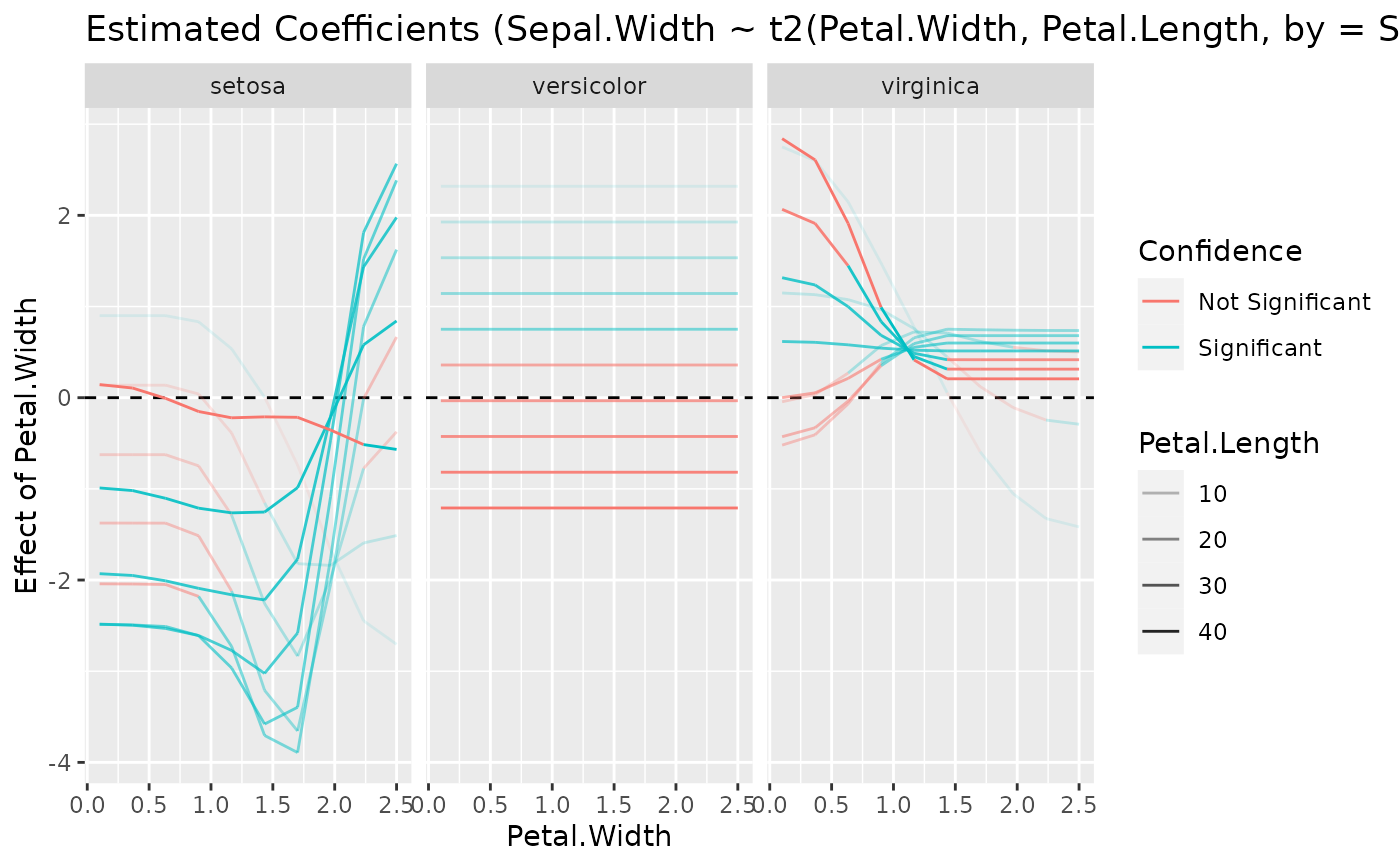
 model <- lm(Petal.Length ~ poly(Sepal.Width, 4), data = iris)
x <- estimate_slopes(model, trend = "Sepal.Width", by = "Sepal.Width", length = 20)
plot(visualisation_recipe(x))
model <- lm(Petal.Length ~ poly(Sepal.Width, 4), data = iris)
x <- estimate_slopes(model, trend = "Sepal.Width", by = "Sepal.Width", length = 20)
plot(visualisation_recipe(x))
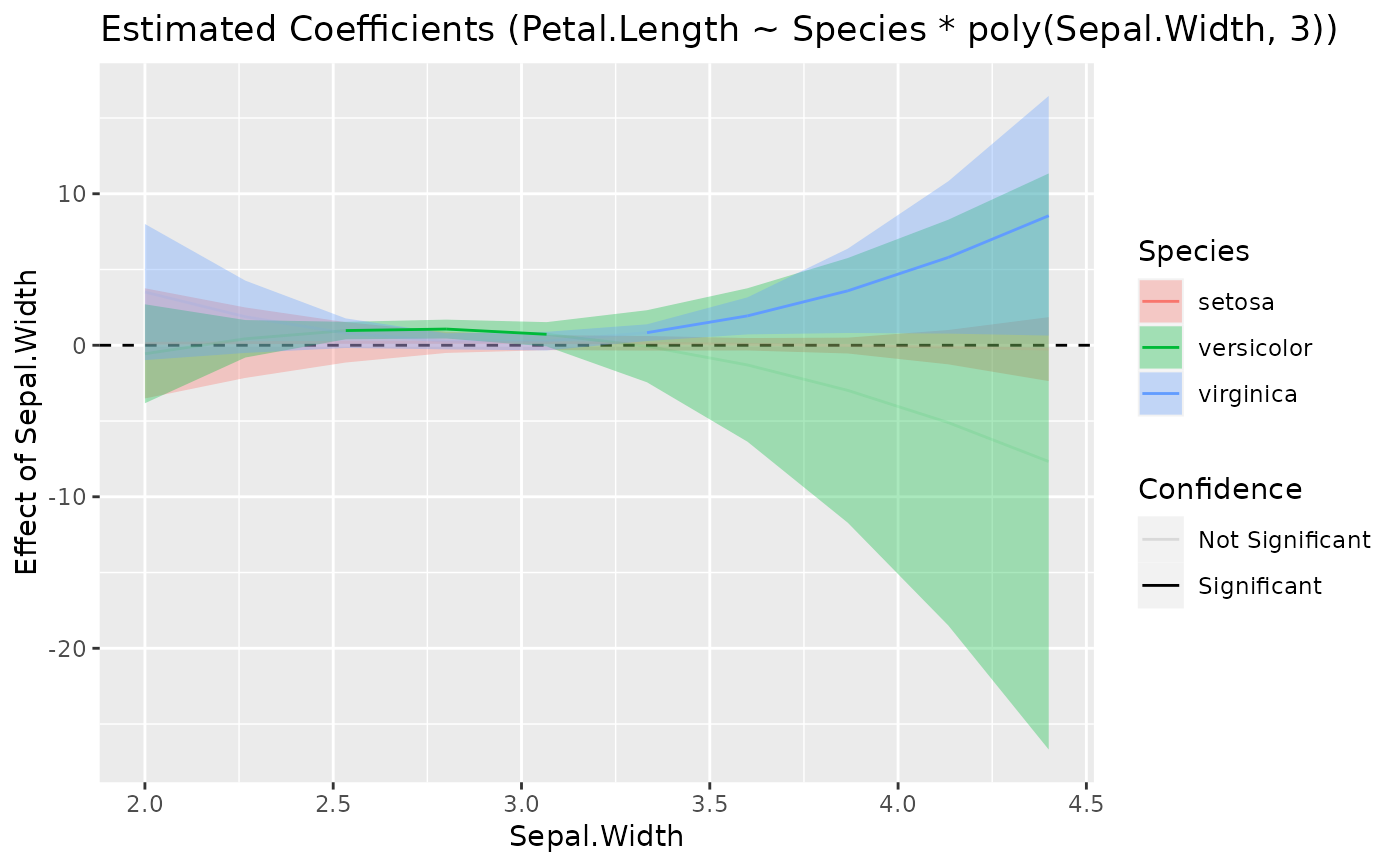
 model <- lm(Petal.Length ~ Species * poly(Sepal.Width, 3), data = iris)
x <- estimate_slopes(model, trend = "Sepal.Width", by = c("Sepal.Width", "Species"))
plot(visualisation_recipe(x))
model <- lm(Petal.Length ~ Species * poly(Sepal.Width, 3), data = iris)
x <- estimate_slopes(model, trend = "Sepal.Width", by = c("Sepal.Width", "Species"))
plot(visualisation_recipe(x))
 # }
# ==============================================
# estimate_grouplevel
# ==============================================
# \dontrun{
data <- lme4::sleepstudy
data <- rbind(data, data)
data$Newfactor <- rep(c("A", "B", "C", "D"))
# 1 random intercept
model <- lme4::lmer(Reaction ~ Days + (1 | Subject), data = data)
x <- estimate_grouplevel(model)
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: pointrange
#> data = [18 x 9]
#> aes_string(
#> y = 'Coefficient'
#> x = 'Level'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#>
#> Layer 2
#> --------
#> Geom type: coord_flip
#>
plot(layers)
# }
# ==============================================
# estimate_grouplevel
# ==============================================
# \dontrun{
data <- lme4::sleepstudy
data <- rbind(data, data)
data$Newfactor <- rep(c("A", "B", "C", "D"))
# 1 random intercept
model <- lme4::lmer(Reaction ~ Days + (1 | Subject), data = data)
x <- estimate_grouplevel(model)
layers <- visualisation_recipe(x)
layers
#> Layer 1
#> --------
#> Geom type: pointrange
#> data = [18 x 9]
#> aes_string(
#> y = 'Coefficient'
#> x = 'Level'
#> ymin = 'CI_low'
#> ymax = 'CI_high'
#> group = '.group'
#> )
#>
#> Layer 2
#> --------
#> Geom type: coord_flip
#>
plot(layers)
 # 2 random intercepts
model <- lme4::lmer(Reaction ~ Days + (1 | Subject) + (1 | Newfactor), data = data)
x <- estimate_grouplevel(model)
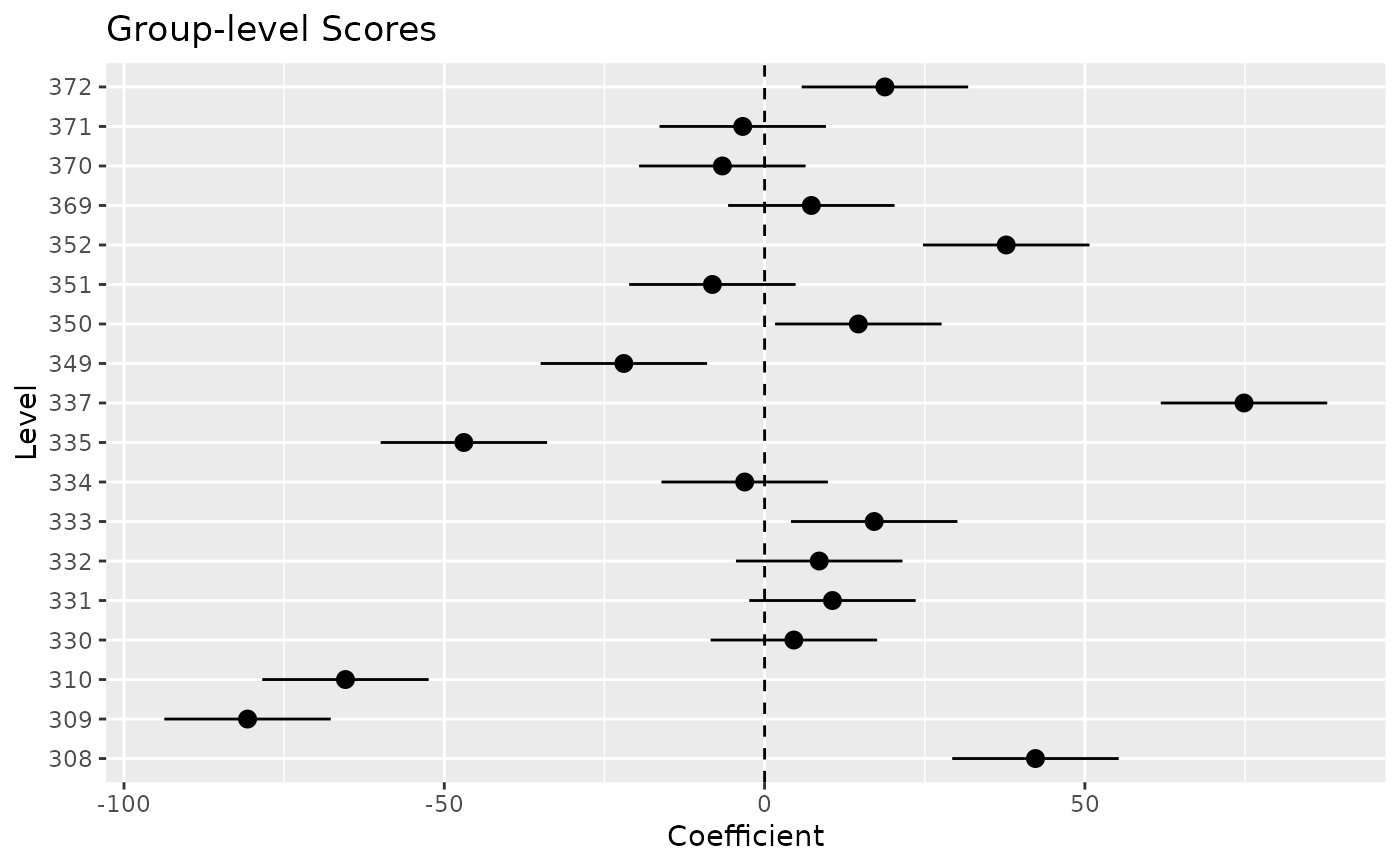
plot(x) +
geom_hline(yintercept = 0, linetype = "dashed") +
theme_minimal()
# 2 random intercepts
model <- lme4::lmer(Reaction ~ Days + (1 | Subject) + (1 | Newfactor), data = data)
x <- estimate_grouplevel(model)
plot(x) +
geom_hline(yintercept = 0, linetype = "dashed") +
theme_minimal()
 # Note: we need to use hline instead of vline because the axes is flipped
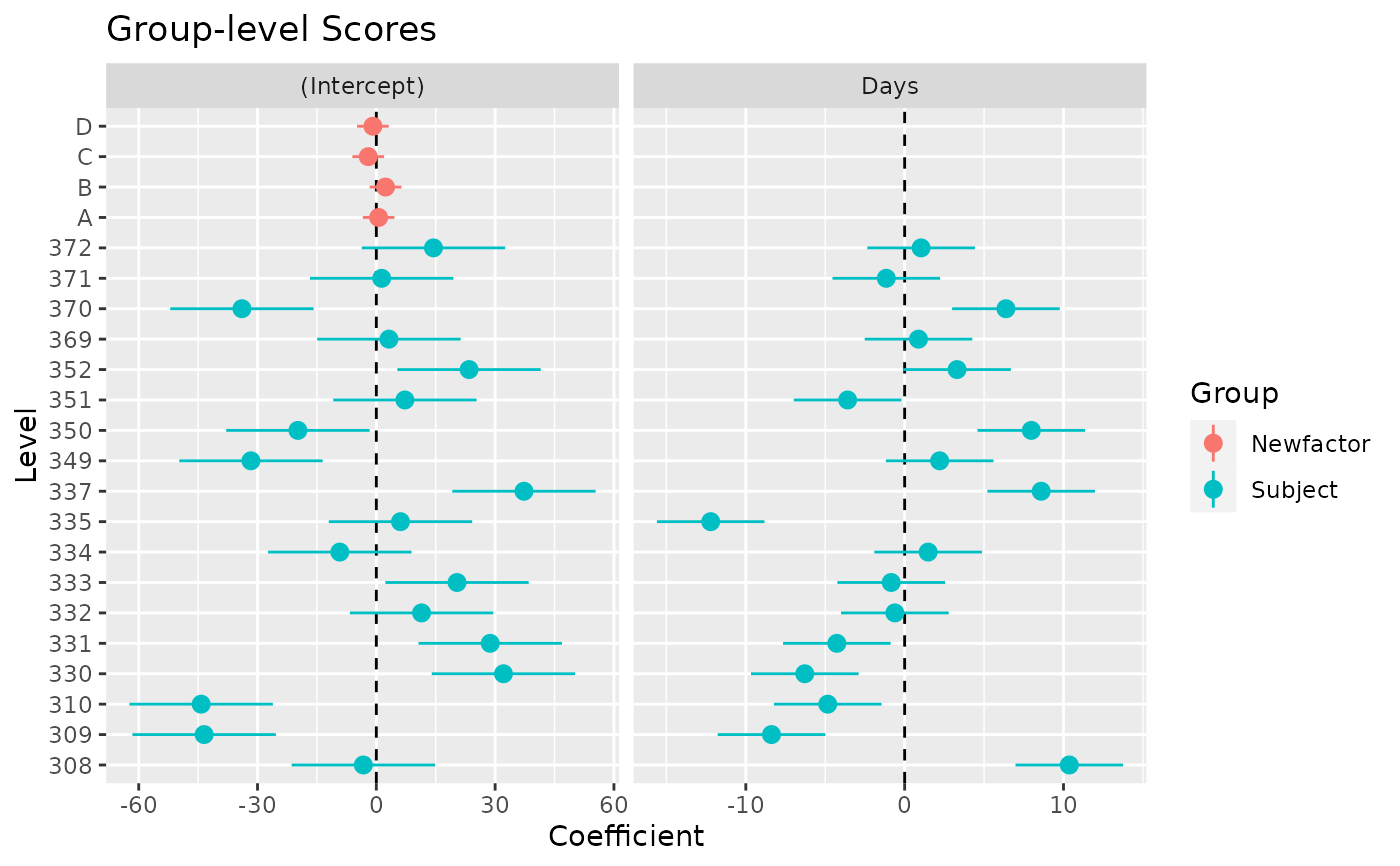
model <- lme4::lmer(Reaction ~ Days + (1 + Days | Subject) + (1 | Newfactor), data = data)
x <- estimate_grouplevel(model)
plot(x)
# Note: we need to use hline instead of vline because the axes is flipped
model <- lme4::lmer(Reaction ~ Days + (1 + Days | Subject) + (1 | Newfactor), data = data)
x <- estimate_grouplevel(model)
plot(x)
 # }
# }