Create a reference matrix, useful for visualisation, with evenly spread and
combined values. Usually used to generate predictions using get_predicted().
See this
vignette
for a tutorial on how to create a visualisation matrix using this function.
Alternatively, these can also be used to extract the "grid" columns from objects generated by emmeans and marginaleffects (see those methods for more info).
Usage
get_datagrid(x, ...)
# S3 method for class 'data.frame'
get_datagrid(
x,
by = "all",
factors = "reference",
numerics = "mean",
length = 10,
range = "range",
preserve_range = FALSE,
protect_integers = TRUE,
digits = 3,
reference = x,
...
)
# S3 method for class 'numeric'
get_datagrid(
x,
length = 10,
range = "range",
protect_integers = TRUE,
digits = 3,
...
)
# S3 method for class 'factor'
get_datagrid(x, ...)
# Default S3 method
get_datagrid(
x,
by = "all",
factors = "reference",
numerics = "mean",
preserve_range = TRUE,
reference = x,
include_smooth = TRUE,
include_random = FALSE,
include_response = FALSE,
data = NULL,
digits = 3,
verbose = TRUE,
...
)Arguments
- x
An object from which to construct the reference grid.
- ...
Arguments passed to or from other methods (for instance,
lengthorrangeto control the spread of numeric variables.).- by
Indicates the focal predictors (variables) for the reference grid and at which values focal predictors should be represented. If not specified otherwise, representative values for numeric variables or predictors are evenly distributed from the minimum to the maximum, with a total number of
lengthvalues covering that range (see 'Examples'). Possible options forbyare:Select variables only:
"all", which will include all variables or predictors.a character vector of one or more variable or predictor names, like
c("Species", "Sepal.Width"), which will create a grid of all combinations of unique values.
Note: If
byspecifies only variable names, without associated values, the following occurs: factor variables use all their levels, numeric variables use a range oflengthequally spaced values between their minimum and maximum, and character variables use all their unique values.Select variables and values:
bycan be a list of named elements, indicating focal predictors and their representative values, e.g.by = list(mpg = 10:20),by = list(Sepal.Length = c(2, 4), Species = "setosa"), orby = list(Sepal.Length = seq(2, 5, 0.5)).Instead of a list, it is possible to write a string representation, or a character vector of such strings, e.g.
by = "mpg = 10:20",by = c("Sepal.Length = c(2, 4)", "Species = 'setosa'"), orby = "Sepal.Length = seq(2, 5, 0.5)". Note the usage of single and double quotes to assign strings within strings.In general, any expression after a
=will be evaluated as R code, which allows using own functions, e.g.
Note: If
byspecifies variables with their associated values, argumentlengthis ignored.
There is a special handling of assignments with brackets, i.e. values defined inside
[and], which create summaries for numeric variables. Following "tokens" that creates pre-defined representative values are possible:for mean and -/+ 1 SD around the mean:
"x = [sd]"for median and -/+ 1 MAD around the median:
"x = [mad]"for Tukey's five number summary (minimum, lower-hinge, median, upper-hinge, maximum):
"x = [fivenum]"for quartiles:
"x = [quartiles]"(same as"x = [fivenum]", but excluding minimum and maximum)for terciles:
"x = [terciles]"for terciles, including minimum and maximum:
"x = [terciles2]"for a pretty value range:
"x = [pretty]"for minimum and maximum value:
"x = [minmax]"for 0 and the maximum value:
"x = [zeromax]"for a random sample from all values:
"x = [sample <number>]", where<number>should be a positive integer, e.g."x = [sample 15]".
Note: the
lengthargument will be ignored when using brackets-tokens.The remaining variables not specified in
bywill be fixed (see also argumentsfactorsandnumerics).- factors
Type of summary for factors not specified in
by. Can be"reference"(set at the reference level),"mode"(set at the most common level) or"all"to keep all levels.- numerics
Type of summary for numeric values not specified in
by. Can be"all"(will duplicate the grid for all unique values), any function ("mean","median", ...) or a value (e.g.,numerics = 0). Special functions are"mode", which will set the numeric variable to its most common value, and"integer", which returns the rounded mean.- length
Length of numeric target variables selected in
by(if no representative values are additionally specified). This arguments controls the number of (equally spread) values that will be taken to represent the continuous (non-integer alike!) variables. A longer length will increase precision, but can also substantially increase the size of the datagrid (especially in case of interactions). IfNA, will return all the unique values.In case of multiple continuous target variables,
lengthcan also be a vector of different values (see 'Examples'). In this case,lengthmust be of same length as numeric target variables. Iflengthis a named vector, values are matched against the names of the target variables.When
range = "range"(the default),lengthis ignored for integer type variables whenlengthis larger than the number of unique values andprotect_integersisTRUE(default). Setprotect_integers = FALSEto create a spread oflengthnumber of values from minimum to maximum for integers, including fractions (i.e., to treat integer variables as regular numeric variables).lengthis furthermore ignored if "tokens" (in brackets[and]) are used inby, or if representative values are additionally specified inby.- range
Option to control the representative values given in
by, if no specific values were provided. Use in combination with thelengthargument to control the number of values within the specified range.rangecan be one of the following:"range"(default), will use the minimum and maximum of the original data vector as end-points (min and max). For integer variables, thelengthargument will be ignored, and"range"will only use values that appear in the data. Setprotect_integers = FALSEto override this behaviour for integer variables.if an interval type is specified, such as
"iqr","ci","hdi"or"eti", it will spread the values within that range (the default CI width is95%but this can be changed by adding for instanceci = 0.90.) SeeIQR()andbayestestR::ci(). This can be useful to have more robust change and skipping extreme values.if
"sd"or"mad", it will spread by this dispersion index around the mean or the median, respectively. If thelengthargument is an even number (e.g.,4), it will have one more step on the positive side (i.e.,-1, 0, +1, +2). The result is a named vector. See 'Examples.'"grid"will create a reference grid that is useful when plotting predictions, by choosing representative values for numeric variables based on their position in the reference grid. If a numeric variable is the first predictor inby, values from minimum to maximum of the same length as indicated inlengthare generated. For numeric predictors not specified at first inby, mean and -1/+1 SD around the mean are returned. For factors, all levels are returned."pretty"will create a range "pretty" values, usingpretty(), where the value inlengthis used for thenargument inpretty().
rangecan also be a vector of different values (see 'Examples'). In this case,rangemust be of same length as numeric target variables. Ifrangeis a named vector, values are matched against the names of the target variables.- preserve_range
In the case of combinations between numeric variables and factors, setting
preserve_range = TRUEwill drop the observations where the value of the numeric variable is originally not present in the range of its factor level. This leads to an unbalanced grid. Also, if you want the minimum and the maximum to closely match the actual ranges, you should increase thelengthargument.- protect_integers
Defaults to
TRUE. Indicates whether integers (whole numbers) should be treated as integers (i.e., prevent adding any in-between round number values), or - ifFALSE- as regular numeric variables. Only applies to focal predictors (specified inby) and when:range = "range"(the default), or ifrange = "grid"and the first predictor inbyis an integer;lengthis larger than the number of unique values for the variable.
If
lengthis smaller than the number of unique values,protect_integersis ignored.- digits
Number of digits used for rounding numeric values specified in
by. E.g.,x = [sd]will round the mean and +-/1 SD in the data grid todigits.- reference
The reference vector from which to compute the mean and SD. Used when standardizing or unstandardizing the grid using
effectsize::standardize.- include_smooth
If
xis a model object, decide whether smooth terms should be included in the data grid or not.- include_random
If
xis a mixed model object, decide whether random effect terms should be included in the data grid or not. Ifinclude_randomisFALSE, butxis a mixed model with random effects, these will still be included in the returned grid, but set to their "population level" value (e.g.,NAfor glmmTMB or0for merMod). This ensures that commonpredict()methods work properly, as these usually need data with all variables in the model included.- include_response
If
xis a model object, decide whether the response variable should be included in the data grid or not.- data
Optional, the data frame that was used to fit the model. Usually, the data is retrieved via
get_data().- verbose
Toggle warnings.
Details
Data grids are an (artificial or theoretical) representation of the sample.
They consists of predictors of interest (so-called focal predictors), and
meaningful values, at which the sample characteristics (focal predictors)
should be represented. The focal predictors are selected in by. To select
meaningful (or representative) values, either use by, or use a combination
of the arguments length and range.
See also
get_predicted() to extract predictions, for which the data grid
is useful, and see the methods for objects generated
by emmeans and marginaleffects to extract the "grid" columns.
Examples
# Datagrids of variables and dataframes =====================================
data(iris)
data(mtcars)
# Single variable is of interest; all others are "fixed" ------------------
# Factors, returns all the levels
get_datagrid(iris, by = "Species")
#> Species Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 1 setosa 5.843333 3.057333 3.758 1.199333
#> 2 versicolor 5.843333 3.057333 3.758 1.199333
#> 3 virginica 5.843333 3.057333 3.758 1.199333
# Specify an expression
get_datagrid(iris, by = "Species = c('setosa', 'versicolor')")
#> Species Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 1 setosa 5.843333 3.057333 3.758 1.199333
#> 2 versicolor 5.843333 3.057333 3.758 1.199333
# Numeric variables, default spread length = 10
get_datagrid(iris, by = "Sepal.Length")
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.3 3.057333 3.758 1.199333 setosa
#> 2 4.7 3.057333 3.758 1.199333 setosa
#> 3 5.1 3.057333 3.758 1.199333 setosa
#> 4 5.5 3.057333 3.758 1.199333 setosa
#> 5 5.9 3.057333 3.758 1.199333 setosa
#> 6 6.3 3.057333 3.758 1.199333 setosa
#> 7 6.7 3.057333 3.758 1.199333 setosa
#> 8 7.1 3.057333 3.758 1.199333 setosa
#> 9 7.5 3.057333 3.758 1.199333 setosa
#> 10 7.9 3.057333 3.758 1.199333 setosa
# change length
get_datagrid(iris, by = "Sepal.Length", length = 3)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.3 3.057333 3.758 1.199333 setosa
#> 2 6.1 3.057333 3.758 1.199333 setosa
#> 3 7.9 3.057333 3.758 1.199333 setosa
# change non-targets fixing
get_datagrid(iris[2:150, ],
by = "Sepal.Length",
factors = "mode", numerics = "median"
)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.3 3 4.4 1.3 versicolor
#> 2 4.7 3 4.4 1.3 versicolor
#> 3 5.1 3 4.4 1.3 versicolor
#> 4 5.5 3 4.4 1.3 versicolor
#> 5 5.9 3 4.4 1.3 versicolor
#> 6 6.3 3 4.4 1.3 versicolor
#> 7 6.7 3 4.4 1.3 versicolor
#> 8 7.1 3 4.4 1.3 versicolor
#> 9 7.5 3 4.4 1.3 versicolor
#> 10 7.9 3 4.4 1.3 versicolor
# change min/max of target
get_datagrid(iris, by = "Sepal.Length", range = "ci", ci = 0.90)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.600 3.057333 3.758 1.199333 setosa
#> 2 4.895 3.057333 3.758 1.199333 setosa
#> 3 5.190 3.057333 3.758 1.199333 setosa
#> 4 5.485 3.057333 3.758 1.199333 setosa
#> 5 5.780 3.057333 3.758 1.199333 setosa
#> 6 6.075 3.057333 3.758 1.199333 setosa
#> 7 6.370 3.057333 3.758 1.199333 setosa
#> 8 6.665 3.057333 3.758 1.199333 setosa
#> 9 6.960 3.057333 3.758 1.199333 setosa
#> 10 7.255 3.057333 3.758 1.199333 setosa
# Manually change min/max
get_datagrid(iris, by = "Sepal.Length = c(0, 1)")
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 0 3.057333 3.758 1.199333 setosa
#> 2 1 3.057333 3.758 1.199333 setosa
# -1 SD, mean and +1 SD
get_datagrid(iris, by = "Sepal.Length = [sd]")
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.015 3.057333 3.758 1.199333 setosa
#> 2 5.843 3.057333 3.758 1.199333 setosa
#> 3 6.671 3.057333 3.758 1.199333 setosa
# rounded to 1 digit
get_datagrid(iris, by = "Sepal.Length = [sd]", digits = 1)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.0 3.057333 3.758 1.199333 setosa
#> 2 5.8 3.057333 3.758 1.199333 setosa
#> 3 6.7 3.057333 3.758 1.199333 setosa
# identical to previous line: -1 SD, mean and +1 SD
get_datagrid(iris, by = "Sepal.Length", range = "sd", length = 3)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.015 3.057333 3.758 1.199333 setosa
#> 2 5.843 3.057333 3.758 1.199333 setosa
#> 3 6.671 3.057333 3.758 1.199333 setosa
# quartiles
get_datagrid(iris, by = "Sepal.Length = [quartiles]")
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.057333 3.758 1.199333 setosa
#> 2 5.8 3.057333 3.758 1.199333 setosa
#> 3 6.4 3.057333 3.758 1.199333 setosa
# Standardization and unstandardization
data <- get_datagrid(iris, by = "Sepal.Length", range = "sd", length = 3)
# It is a named vector (extract names with `names(out$Sepal.Length)`)
data$Sepal.Length
#> -1 SD Mean +1 SD
#> 5.015 5.843 6.671
datawizard::standardize(data, select = "Sepal.Length")
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 -1.0003226860 3.057333 3.758 1.199333 setosa
#> 2 -0.0004025443 3.057333 3.758 1.199333 setosa
#> 3 0.9995175973 3.057333 3.758 1.199333 setosa
# Manually specify values
data <- get_datagrid(iris, by = "Sepal.Length = c(-2, 0, 2)")
data
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 -2 3.057333 3.758 1.199333 setosa
#> 2 0 3.057333 3.758 1.199333 setosa
#> 3 2 3.057333 3.758 1.199333 setosa
datawizard::unstandardize(data, select = "Sepal.Length")
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.187201 3.057333 3.758 1.199333 setosa
#> 2 5.843333 3.057333 3.758 1.199333 setosa
#> 3 7.499466 3.057333 3.758 1.199333 setosa
# Multiple variables are of interest, creating a combination --------------
get_datagrid(iris, by = c("Sepal.Length", "Species"), length = 3)
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 4.3 setosa 3.057333 3.758 1.199333
#> 2 6.1 setosa 3.057333 3.758 1.199333
#> 3 7.9 setosa 3.057333 3.758 1.199333
#> 4 4.3 versicolor 3.057333 3.758 1.199333
#> 5 6.1 versicolor 3.057333 3.758 1.199333
#> 6 7.9 versicolor 3.057333 3.758 1.199333
#> 7 4.3 virginica 3.057333 3.758 1.199333
#> 8 6.1 virginica 3.057333 3.758 1.199333
#> 9 7.9 virginica 3.057333 3.758 1.199333
get_datagrid(iris, by = c("Sepal.Length", "Petal.Length"), length = c(3, 2))
#> Sepal.Length Petal.Length Sepal.Width Petal.Width Species
#> 1 4.3 1.0 3.057333 1.199333 setosa
#> 2 6.1 1.0 3.057333 1.199333 setosa
#> 3 7.9 1.0 3.057333 1.199333 setosa
#> 4 4.3 6.9 3.057333 1.199333 setosa
#> 5 6.1 6.9 3.057333 1.199333 setosa
#> 6 7.9 6.9 3.057333 1.199333 setosa
get_datagrid(iris, by = c(1, 3), length = 3)
#> Sepal.Length Petal.Length Sepal.Width Petal.Width Species
#> 1 4.3 1.00 3.057333 1.199333 setosa
#> 2 6.1 1.00 3.057333 1.199333 setosa
#> 3 7.9 1.00 3.057333 1.199333 setosa
#> 4 4.3 3.95 3.057333 1.199333 setosa
#> 5 6.1 3.95 3.057333 1.199333 setosa
#> 6 7.9 3.95 3.057333 1.199333 setosa
#> 7 4.3 6.90 3.057333 1.199333 setosa
#> 8 6.1 6.90 3.057333 1.199333 setosa
#> 9 7.9 6.90 3.057333 1.199333 setosa
get_datagrid(iris, by = c("Sepal.Length", "Species"), preserve_range = TRUE)
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 4.3 setosa 3.057333 3.758 1.199333
#> 2 4.7 setosa 3.057333 3.758 1.199333
#> 3 5.1 setosa 3.057333 3.758 1.199333
#> 4 5.5 setosa 3.057333 3.758 1.199333
#> 5 5.1 versicolor 3.057333 3.758 1.199333
#> 6 5.5 versicolor 3.057333 3.758 1.199333
#> 7 5.9 versicolor 3.057333 3.758 1.199333
#> 8 6.3 versicolor 3.057333 3.758 1.199333
#> 9 6.7 versicolor 3.057333 3.758 1.199333
#> 10 5.1 virginica 3.057333 3.758 1.199333
#> 11 5.5 virginica 3.057333 3.758 1.199333
#> 12 5.9 virginica 3.057333 3.758 1.199333
#> 13 6.3 virginica 3.057333 3.758 1.199333
#> 14 6.7 virginica 3.057333 3.758 1.199333
#> 15 7.1 virginica 3.057333 3.758 1.199333
#> 16 7.5 virginica 3.057333 3.758 1.199333
#> 17 7.9 virginica 3.057333 3.758 1.199333
get_datagrid(iris, by = c("Sepal.Length", "Species"), numerics = 0)
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 4.3 setosa 0 0 0
#> 2 4.7 setosa 0 0 0
#> 3 5.1 setosa 0 0 0
#> 4 5.5 setosa 0 0 0
#> 5 5.9 setosa 0 0 0
#> 6 6.3 setosa 0 0 0
#> 7 6.7 setosa 0 0 0
#> 8 7.1 setosa 0 0 0
#> 9 7.5 setosa 0 0 0
#> 10 7.9 setosa 0 0 0
#> 11 4.3 versicolor 0 0 0
#> 12 4.7 versicolor 0 0 0
#> 13 5.1 versicolor 0 0 0
#> 14 5.5 versicolor 0 0 0
#> 15 5.9 versicolor 0 0 0
#> 16 6.3 versicolor 0 0 0
#> 17 6.7 versicolor 0 0 0
#> 18 7.1 versicolor 0 0 0
#> 19 7.5 versicolor 0 0 0
#> 20 7.9 versicolor 0 0 0
#> 21 4.3 virginica 0 0 0
#> 22 4.7 virginica 0 0 0
#> 23 5.1 virginica 0 0 0
#> 24 5.5 virginica 0 0 0
#> 25 5.9 virginica 0 0 0
#> 26 6.3 virginica 0 0 0
#> 27 6.7 virginica 0 0 0
#> 28 7.1 virginica 0 0 0
#> 29 7.5 virginica 0 0 0
#> 30 7.9 virginica 0 0 0
get_datagrid(iris, by = c("Sepal.Length = 3", "Species"))
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 3 setosa 3.057333 3.758 1.199333
#> 2 3 versicolor 3.057333 3.758 1.199333
#> 3 3 virginica 3.057333 3.758 1.199333
get_datagrid(iris, by = c("Sepal.Length = c(3, 1)", "Species = 'setosa'"))
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 3 setosa 3.057333 3.758 1.199333
#> 2 1 setosa 3.057333 3.758 1.199333
# specify length individually for each focal predictor
# values are matched by names
get_datagrid(mtcars[1:4], by = c("mpg", "hp"), length = c(hp = 3, mpg = 2))
#> mpg hp cyl disp
#> 1 10.4 52.0 6.1875 230.7219
#> 2 33.9 52.0 6.1875 230.7219
#> 3 10.4 193.5 6.1875 230.7219
#> 4 33.9 193.5 6.1875 230.7219
#> 5 10.4 335.0 6.1875 230.7219
#> 6 33.9 335.0 6.1875 230.7219
# Numeric and categorical variables, generating a grid for plots
# default spread when numerics are first: length = 10
get_datagrid(iris, by = c("Sepal.Length", "Species"), range = "grid")
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 4.3 setosa 3.057333 3.758 1.199333
#> 2 4.7 setosa 3.057333 3.758 1.199333
#> 3 5.1 setosa 3.057333 3.758 1.199333
#> 4 5.5 setosa 3.057333 3.758 1.199333
#> 5 5.9 setosa 3.057333 3.758 1.199333
#> 6 6.3 setosa 3.057333 3.758 1.199333
#> 7 6.7 setosa 3.057333 3.758 1.199333
#> 8 7.1 setosa 3.057333 3.758 1.199333
#> 9 7.5 setosa 3.057333 3.758 1.199333
#> 10 7.9 setosa 3.057333 3.758 1.199333
#> 11 4.3 versicolor 3.057333 3.758 1.199333
#> 12 4.7 versicolor 3.057333 3.758 1.199333
#> 13 5.1 versicolor 3.057333 3.758 1.199333
#> 14 5.5 versicolor 3.057333 3.758 1.199333
#> 15 5.9 versicolor 3.057333 3.758 1.199333
#> 16 6.3 versicolor 3.057333 3.758 1.199333
#> 17 6.7 versicolor 3.057333 3.758 1.199333
#> 18 7.1 versicolor 3.057333 3.758 1.199333
#> 19 7.5 versicolor 3.057333 3.758 1.199333
#> 20 7.9 versicolor 3.057333 3.758 1.199333
#> 21 4.3 virginica 3.057333 3.758 1.199333
#> 22 4.7 virginica 3.057333 3.758 1.199333
#> 23 5.1 virginica 3.057333 3.758 1.199333
#> 24 5.5 virginica 3.057333 3.758 1.199333
#> 25 5.9 virginica 3.057333 3.758 1.199333
#> 26 6.3 virginica 3.057333 3.758 1.199333
#> 27 6.7 virginica 3.057333 3.758 1.199333
#> 28 7.1 virginica 3.057333 3.758 1.199333
#> 29 7.5 virginica 3.057333 3.758 1.199333
#> 30 7.9 virginica 3.057333 3.758 1.199333
# default spread when numerics are not first: length = 3 (-1 SD, mean and +1 SD)
get_datagrid(iris, by = c("Species", "Sepal.Length"), range = "grid")
#> Species Sepal.Length Sepal.Width Petal.Length Petal.Width
#> 1 setosa 5.015 3.057333 3.758 1.199333
#> 2 setosa 5.843 3.057333 3.758 1.199333
#> 3 setosa 6.671 3.057333 3.758 1.199333
#> 4 versicolor 5.015 3.057333 3.758 1.199333
#> 5 versicolor 5.843 3.057333 3.758 1.199333
#> 6 versicolor 6.671 3.057333 3.758 1.199333
#> 7 virginica 5.015 3.057333 3.758 1.199333
#> 8 virginica 5.843 3.057333 3.758 1.199333
#> 9 virginica 6.671 3.057333 3.758 1.199333
# range of values
get_datagrid(iris, by = c("Sepal.Width = 1:5", "Petal.Width = 1:3"))
#> Sepal.Width Petal.Width Sepal.Length Petal.Length Species
#> 1 1 1 5.843333 3.758 setosa
#> 2 2 1 5.843333 3.758 setosa
#> 3 3 1 5.843333 3.758 setosa
#> 4 4 1 5.843333 3.758 setosa
#> 5 5 1 5.843333 3.758 setosa
#> 6 1 2 5.843333 3.758 setosa
#> 7 2 2 5.843333 3.758 setosa
#> 8 3 2 5.843333 3.758 setosa
#> 9 4 2 5.843333 3.758 setosa
#> 10 5 2 5.843333 3.758 setosa
#> 11 1 3 5.843333 3.758 setosa
#> 12 2 3 5.843333 3.758 setosa
#> 13 3 3 5.843333 3.758 setosa
#> 14 4 3 5.843333 3.758 setosa
#> 15 5 3 5.843333 3.758 setosa
# With list-style by-argument
get_datagrid(
iris,
by = list(Sepal.Length = 1:3, Species = c("setosa", "versicolor"))
)
#> Sepal.Length Species Sepal.Width Petal.Length Petal.Width
#> 1 1 setosa 3.057333 3.758 1.199333
#> 2 2 setosa 3.057333 3.758 1.199333
#> 3 3 setosa 3.057333 3.758 1.199333
#> 4 1 versicolor 3.057333 3.758 1.199333
#> 5 2 versicolor 3.057333 3.758 1.199333
#> 6 3 versicolor 3.057333 3.758 1.199333
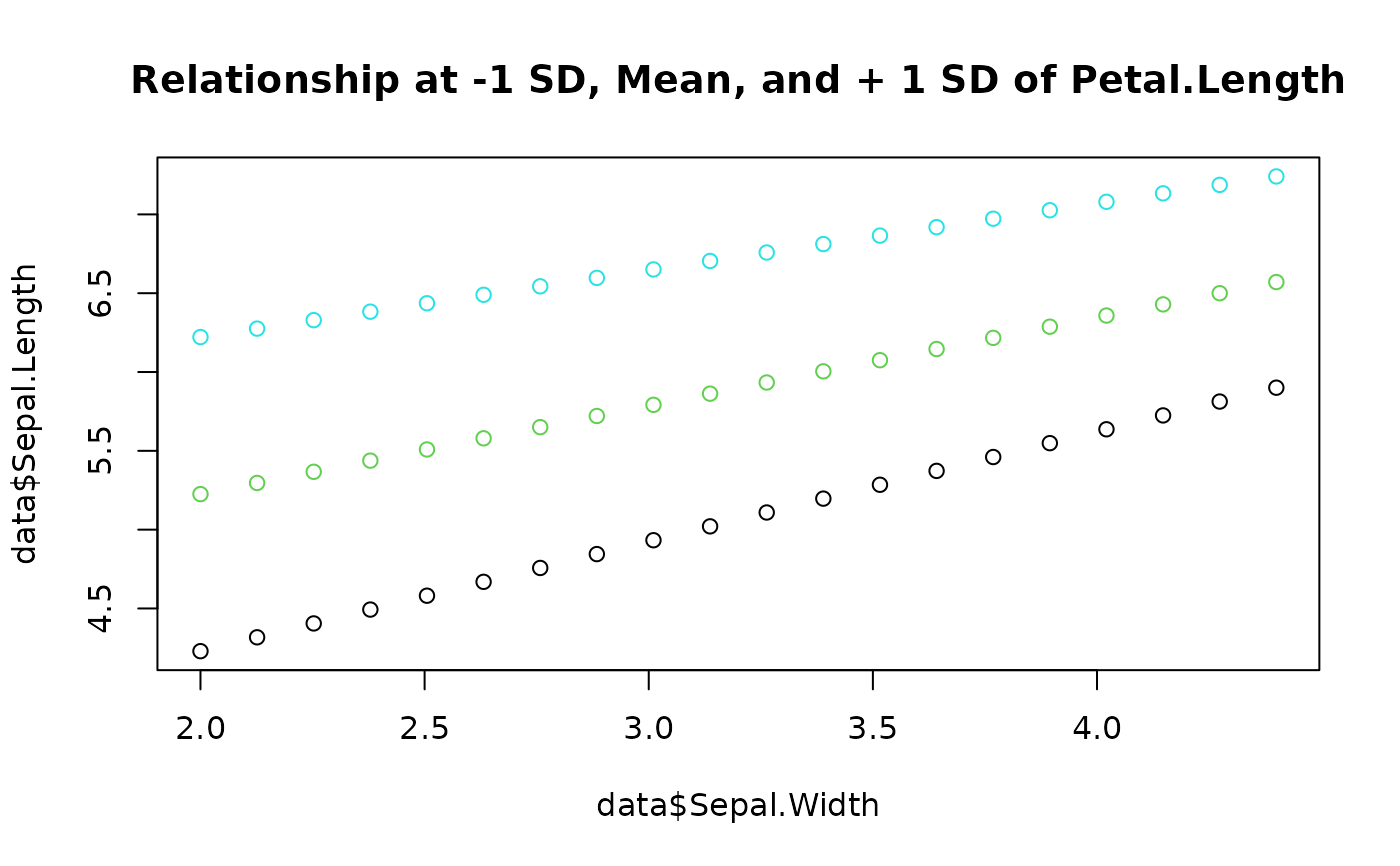
# With models ===============================================================
# Fit a linear regression
model <- lm(Sepal.Length ~ Sepal.Width * Petal.Length, data = iris)
# Get datagrid of predictors
data <- get_datagrid(model, length = c(20, 3), range = c("range", "sd"))
# same as: get_datagrid(model, range = "grid", length = 20)
# Add predictions
data$Sepal.Length <- get_predicted(model, data = data)
# Visualize relationships (each color is at -1 SD, Mean, and + 1 SD of Petal.Length)
plot(data$Sepal.Width, data$Sepal.Length,
col = data$Petal.Length,
main = "Relationship at -1 SD, Mean, and + 1 SD of Petal.Length"
)