Plot method for (conditional) equivalence testing
Source:R/plot.effectsize_table.R, R/plot.equivalence_test.R
plot.see_equivalence_test.RdThe plot() method for the bayestestR::equivalence_test() function.
Usage
# S3 method for class 'see_equivalence_test_effectsize'
plot(x, ...)
# S3 method for class 'see_equivalence_test'
plot(
x,
color_rope = "#0171D3",
alpha_rope = 0.2,
show_intercept = FALSE,
n_columns = 1,
...
)
# S3 method for class 'see_equivalence_test_lm'
plot(
x,
size_point = 0.7,
color_rope = "#0171D3",
alpha_rope = 0.2,
show_intercept = FALSE,
n_columns = 1,
...
)Arguments
- x
An object.
- ...
Arguments passed to or from other methods.
- color_rope
Character specifying color of ROPE ribbon.
- alpha_rope
Numeric specifying transparency level of ROPE ribbon.
- show_intercept
Logical, if
TRUE, the intercept-parameter is included in the plot. By default, it is hidden because in many cases the intercept-parameter has a posterior distribution on a very different location, so density curves of posterior distributions for other parameters are hardly visible.- n_columns
For models with multiple components (like fixed and random, count and zero-inflated), defines the number of columns for the panel-layout. If
NULL, a single, integrated plot is shown.- size_point
Numeric specifying size of point-geoms.
Examples
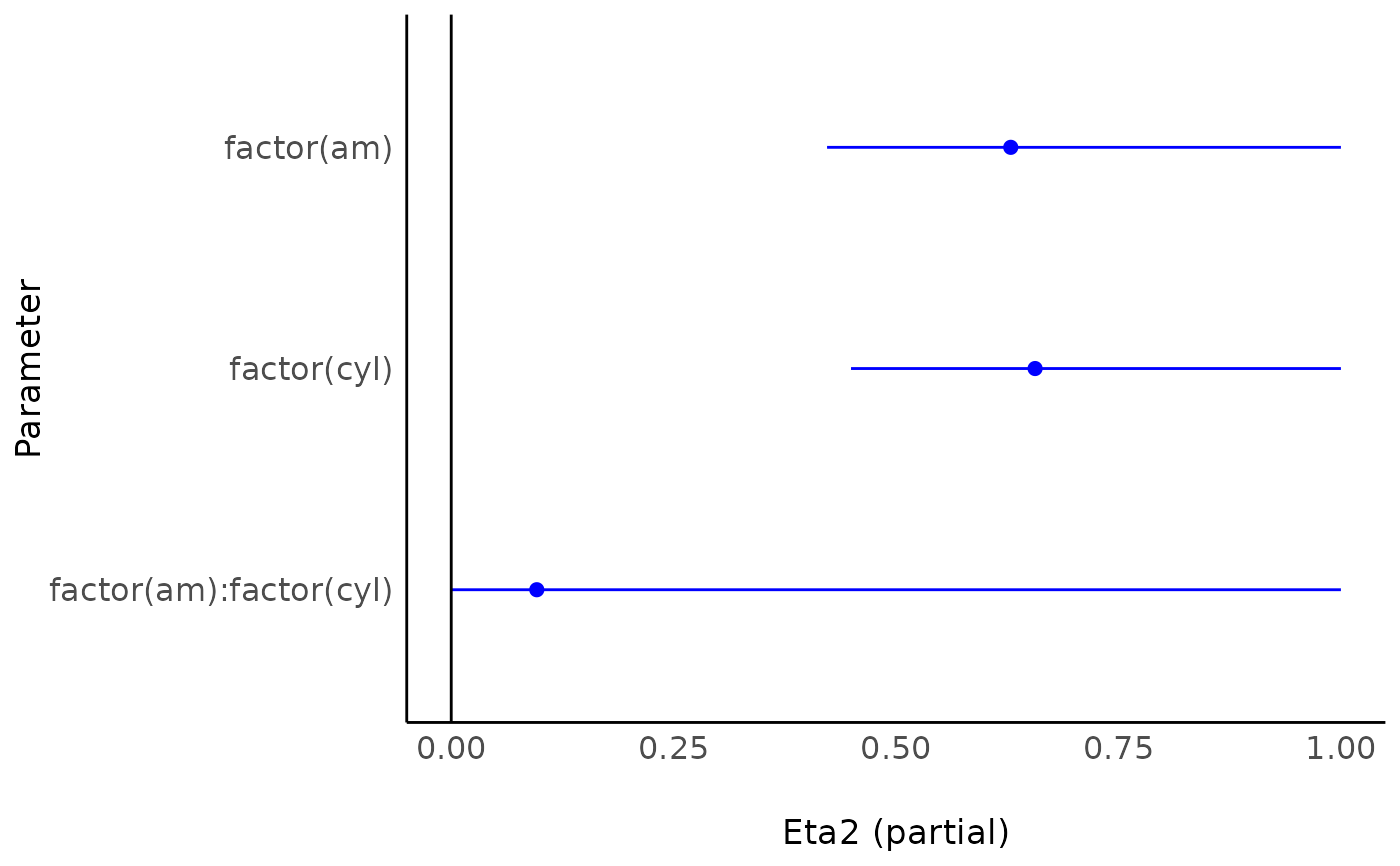
library(effectsize)
m <- aov(mpg ~ factor(am) * factor(cyl), data = mtcars)
result <- eta_squared(m)
plot(result)