For univariate distributions (one-dimensional vectors), this functions performs a Ameijeiras-Alonso et al. (2018) excess mass test. For multivariate distributions (data frames), it uses mixture modelling. However, it seems that it always returns a significant result (suggesting that the distribution is multimodal). A better method might be needed here.
References
Ameijeiras-Alonso, J., Crujeiras, R. M., and Rodríguez-Casal, A. (2019). Mode testing, critical bandwidth and excess mass. Test, 28(3), 900-919.
Examples
# \donttest{
# Univariate
x <- rnorm(1000)
check_multimodal(x)
#> # Is the variable multimodal?
#>
#> The Ameijeiras-Alonso et al. (2018) excess mass test suggests that the
#> hypothesis of a multimodal distribution cannot be rejected (excess mass
#> = 0.02, p = 0.174).
#>
x <- c(rnorm(1000), rnorm(1000, 2))
check_multimodal(x)
#> # Is the variable multimodal?
#>
#> The Ameijeiras-Alonso et al. (2018) excess mass test suggests that the
#> hypothesis of a multimodal distribution cannot be rejected (excess mass
#> = 0.01, p = 0.810).
#>
# Multivariate
m <- data.frame(
x = rnorm(200),
y = rbeta(200, 2, 1)
)
plot(m$x, m$y)
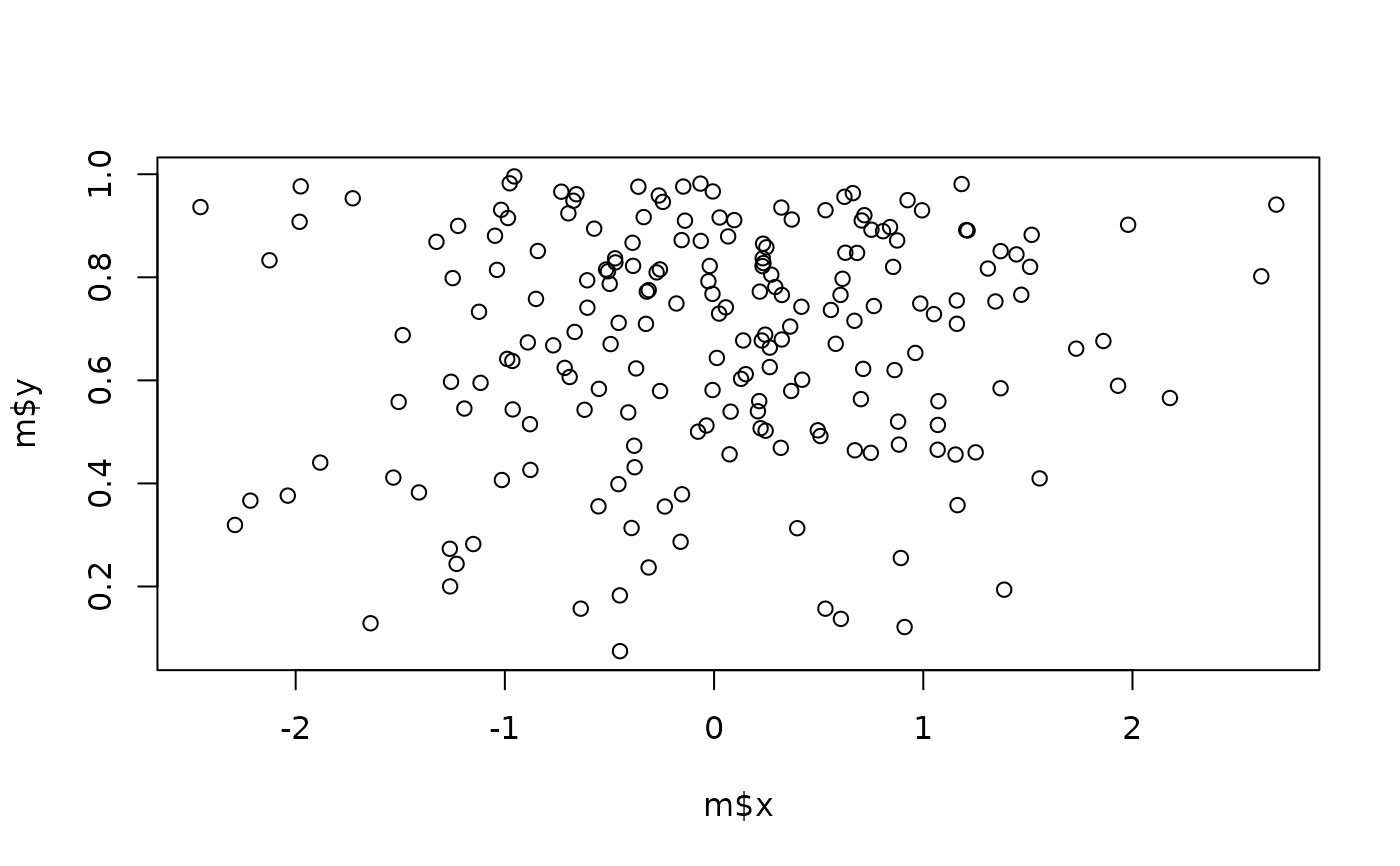 check_multimodal(m)
#> # Is the data multimodal?
#>
#> The parametric mixture modelling test suggests that the multivariate
#> distribution is significantly multimodal (Chi2(7) = 21.34, p = 0.003).
#>
m <- data.frame(
x = c(rnorm(100), rnorm(100, 4)),
y = c(rbeta(100, 2, 1), rbeta(100, 1, 4))
)
plot(m$x, m$y)
check_multimodal(m)
#> # Is the data multimodal?
#>
#> The parametric mixture modelling test suggests that the multivariate
#> distribution is significantly multimodal (Chi2(7) = 21.34, p = 0.003).
#>
m <- data.frame(
x = c(rnorm(100), rnorm(100, 4)),
y = c(rbeta(100, 2, 1), rbeta(100, 1, 4))
)
plot(m$x, m$y)
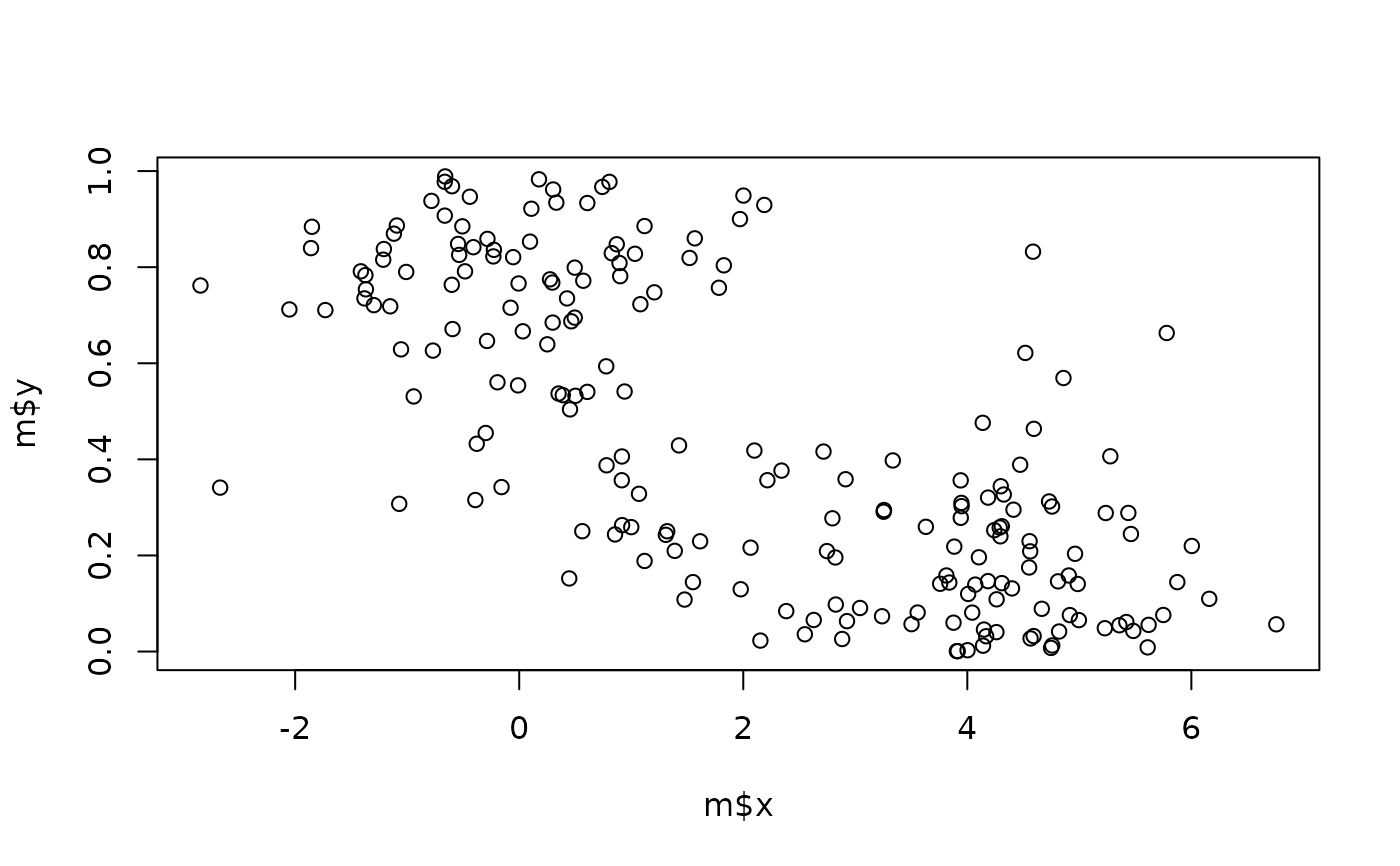 check_multimodal(m)
#> # Is the data multimodal?
#>
#> The parametric mixture modelling test suggests that the multivariate
#> distribution is significantly multimodal (Chi2(11) = 104.48, p < .001).
#>
# }
check_multimodal(m)
#> # Is the data multimodal?
#>
#> The parametric mixture modelling test suggests that the multivariate
#> distribution is significantly multimodal (Chi2(11) = 104.48, p < .001).
#>
# }
