Check model quality of binomial logistic regression models.
Usage
binned_residuals(
model,
term = NULL,
n_bins = NULL,
show_dots = NULL,
ci = 0.95,
ci_type = "exact",
residuals = "deviance",
iterations = 1000,
verbose = TRUE,
...
)Arguments
- model
A
glm-object with binomial-family.- term
Name of independent variable from
x. If notNULL, average residuals for the categories oftermare plotted; else, average residuals for the estimated probabilities of the response are plotted.- n_bins
Numeric, the number of bins to divide the data. If
n_bins = NULL, the square root of the number of observations is taken.- show_dots
Logical, if
TRUE, will show data points in the plot. Set toFALSEfor models with many observations, if generating the plot is too time-consuming. By default,show_dots = NULL. In this casebinned_residuals()tries to guess whether performance will be poor due to a very large model and thus automatically shows or hides dots.- ci
Numeric, the confidence level for the error bounds.
- ci_type
Character, the type of error bounds to calculate. Can be
"exact"(default),"gaussian"or"boot"."exact"calculates the error bounds based on the exact binomial distribution, usingbinom.test()."gaussian"uses the Gaussian approximation, while"boot"uses a simple bootstrap method, where confidence intervals are calculated based on the quantiles of the bootstrap distribution.- residuals
Character, the type of residuals to calculate. Can be
"deviance"(default),"pearson"or"response". It is recommended to use"response"only for those models where other residuals are not available.- iterations
Integer, the number of iterations to use for the bootstrap method. Only used if
ci_type = "boot".- verbose
Toggle warnings and messages.
- ...
Currently not used.
Value
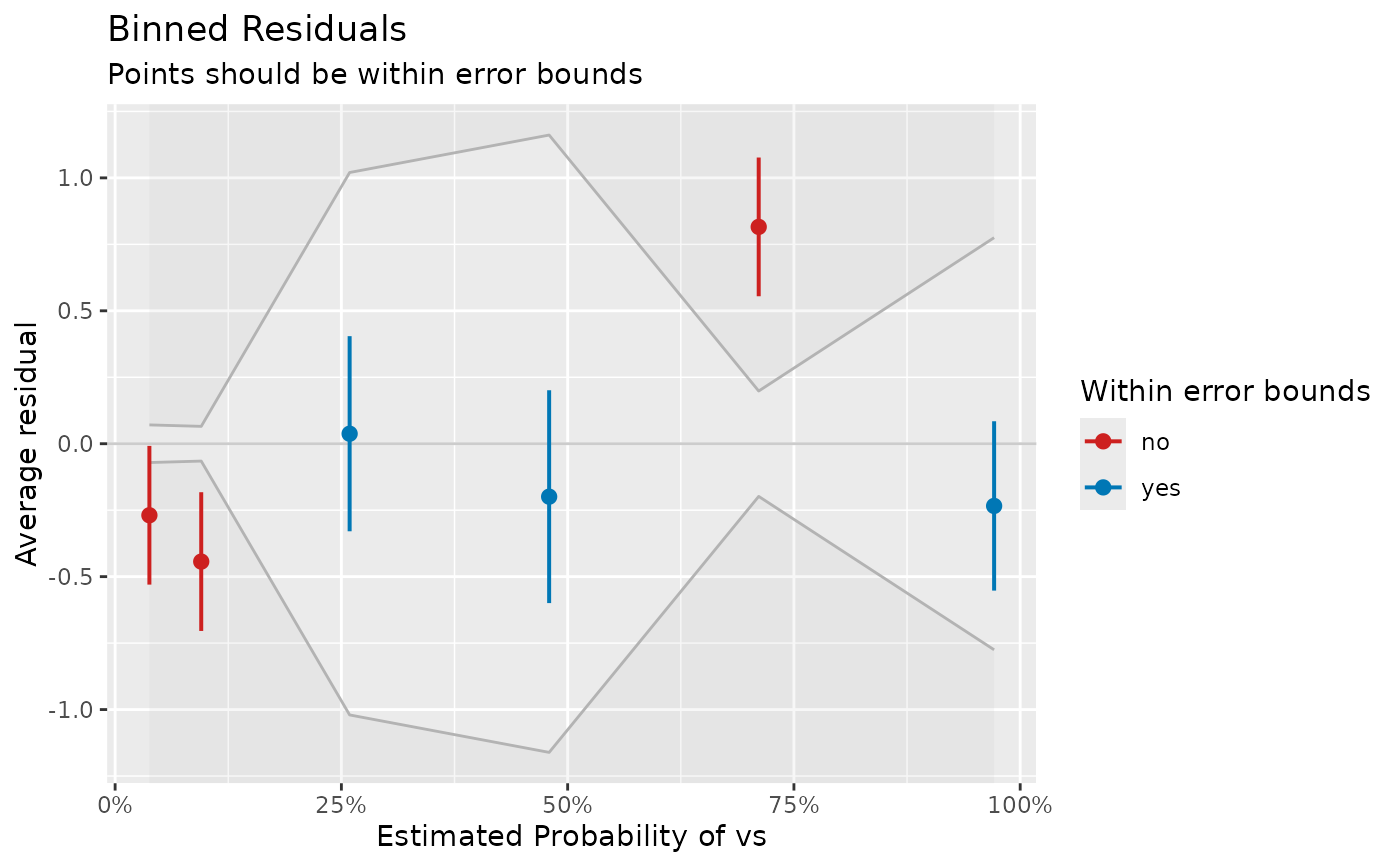
A data frame representing the data that is mapped in the accompanying plot. In case all residuals are inside the error bounds, points are black. If some of the residuals are outside the error bounds (indicated by the grey-shaded area), blue points indicate residuals that are OK, while red points indicate model under- or over-fitting for the relevant range of estimated probabilities.
Details
Binned residual plots are achieved by "dividing the data into categories (bins) based on their fitted values, and then plotting the average residual versus the average fitted value for each bin." (Gelman, Hill 2007: 97). If the model were true, one would expect about 95% of the residuals to fall inside the error bounds.
If term is not NULL, one can compare the residuals in
relation to a specific model predictor. This may be helpful to check if a
term would fit better when transformed, e.g. a rising and falling pattern
of residuals along the x-axis is a signal to consider taking the logarithm
of the predictor (cf. Gelman and Hill 2007, pp. 97-98).
Note
binned_residuals() returns a data frame, however, the print()
method only returns a short summary of the result. The data frame itself
is used for plotting. The plot() method, in turn, creates a ggplot-object.
References
Gelman, A., and Hill, J. (2007). Data analysis using regression and multilevel/hierarchical models. Cambridge; New York: Cambridge University Press.
Examples
model <- glm(vs ~ wt + mpg, data = mtcars, family = "binomial")
result <- binned_residuals(model)
result
#> Warning: Probably bad model fit. Only about 50% of the residuals are inside the error bounds.
#>
# look at the data frame
as.data.frame(result)
#> xbar ybar n x.lo x.hi se CI_low
#> conf_int 0.03786483 -0.26905395 5 0.01744776 0.06917366 0.07079661 -0.5299658
#> conf_int1 0.09514191 -0.44334345 5 0.07087498 0.15160143 0.06530245 -0.7042553
#> conf_int2 0.25910531 0.03762945 6 0.17159955 0.35374001 1.02017708 -0.3293456
#> conf_int3 0.47954643 -0.19916717 5 0.38363314 0.54063600 1.16107852 -0.5994783
#> conf_int4 0.71108931 0.81563262 5 0.57299903 0.89141359 0.19814385 0.5547207
#> conf_int5 0.97119262 -0.23399465 6 0.91147360 0.99815623 0.77513642 -0.5525066
#> CI_high group
#> conf_int -0.008142076 no
#> conf_int1 -0.182431572 no
#> conf_int2 0.404604465 yes
#> conf_int3 0.201143953 yes
#> conf_int4 1.076544495 no
#> conf_int5 0.084517267 yes
# \donttest{
# plot
plot(result, show_dots = TRUE)
 # }
# }
