
Plot method for Model Parameters from Bayesian Meta-Analysis
Source:R/plot.parameters_brms_meta.R
plot.see_parameters_brms_meta.RdThe plot() method for the parameters::model_parameters()
function when used with brms-meta-analysis models.
Usage
# S3 method for class 'see_parameters_brms_meta'
plot(
x,
size_point = 2,
linewidth = 0.8,
size_text = 3.5,
alpha_posteriors = 0.7,
alpha_rope = 0.15,
color_rope = "cadetblue",
normalize_height = TRUE,
show_labels = TRUE,
...
)Arguments
- x
An object.
- size_point
Numeric specifying size of point-geoms.
- linewidth
Numeric value specifying size of line geoms.
- size_text
Numeric value specifying size of text labels.
- alpha_posteriors
Numeric value specifying alpha for the posterior distributions.
- alpha_rope
Numeric specifying transparency level of ROPE ribbon.
- color_rope
Character specifying color of ROPE ribbon.
- normalize_height
Logical. If
TRUE, height of mcmc-areas is "normalized", to avoid overlap. In certain cases when the range of a posterior distribution is narrow for some parameters, this may result in very flat mcmc-areas. In such cases, setnormalize_height = FALSE.- show_labels
Logical. If
TRUE, text labels are displayed.- ...
Arguments passed to or from other methods.
Details
Colors of density areas and errorbars
To change the colors of the density areas, use scale_fill_manual()
with named color-values, e.g. scale_fill_manual(values = c("Study" = "blue", "Overall" = "green")).
To change the color of the error bars, use scale_color_manual(values = c("Errorbar" = "red")).
Examples
# \donttest{
library(parameters)
library(brms)
library(metafor)
data(dat.bcg)
dat <- escalc(
measure = "RR",
ai = tpos,
bi = tneg,
ci = cpos,
di = cneg,
data = dat.bcg
)
dat$author <- make.unique(dat$author)
# model
set.seed(123)
priors <- c(
prior(normal(0, 1), class = Intercept),
prior(cauchy(0, 0.5), class = sd)
)
model <- suppressWarnings(
brm(yi | se(vi) ~ 1 + (1 | author), data = dat, refresh = 0, silent = 2)
)
# result
mp <- model_parameters(model)
plot(mp)
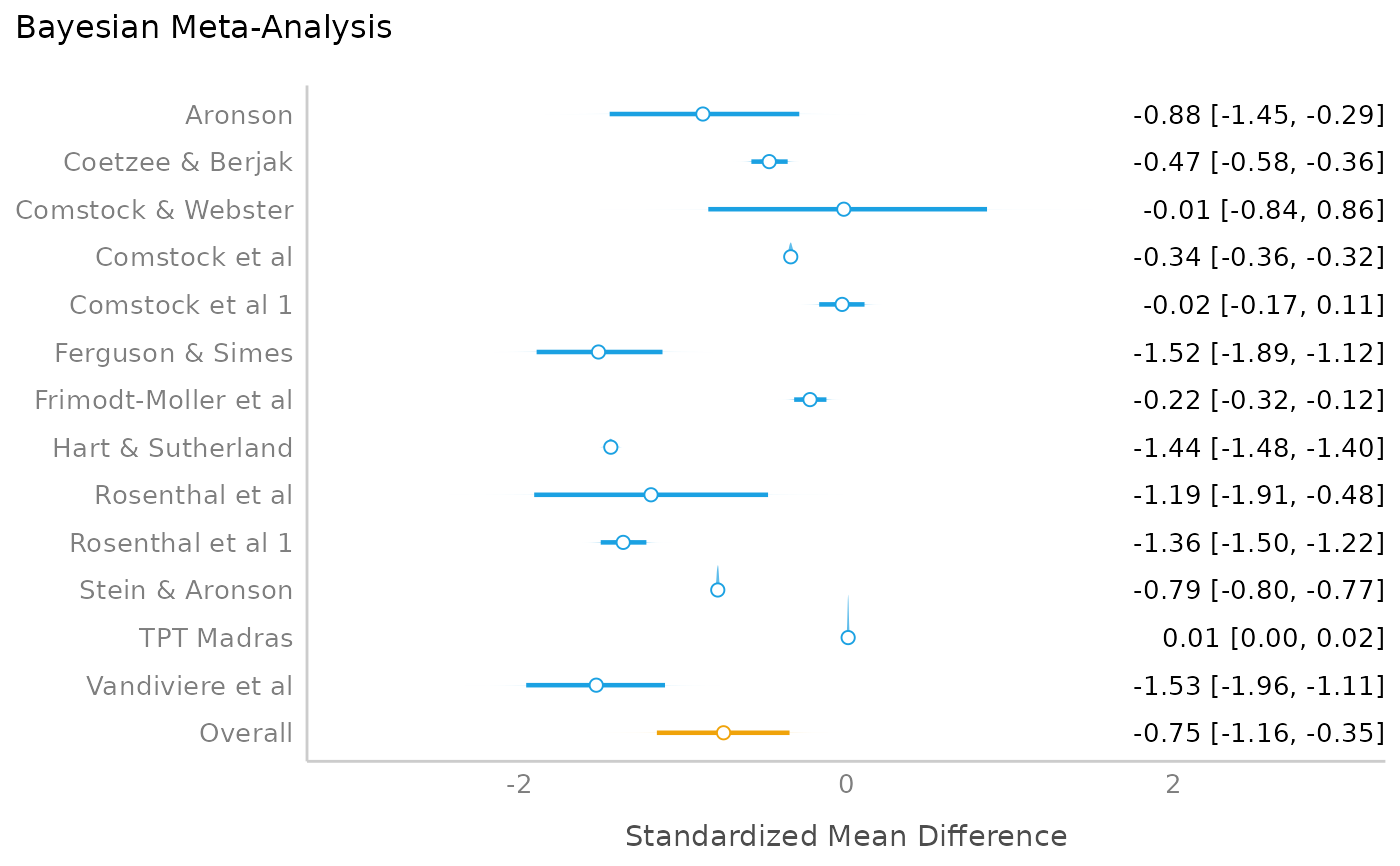 # }
# }