
Okay, “an alternative to broom” might be a bit of an overstatement (at least for now…)
But the parameters package, finally on CRAN, already has some cool features!
parameters
We have recently decided to collaborate around the easystats project, a set of packages designed to make your life easier. This project encompasses several packages, devoted for instance to model internal access, Bayesian analysis, as well as indices of model performance and quality.
parameters' primary goal is to provide utilities for processing the parameters of various statistical models. Beyond computing p-values, CIs, Bayesian indices and other measures for a wide variety of models, this package implements features like standardization or bootstrapping of parameters and models, feature reduction (feature extraction and variable selection) as well as conversion between indices of effect size.
The main function of the package is model_parameters(), which allows you to extract the parameters and their characteristics from various models in a consistent way. It can be considered as a lightweight alternative to broom::tidy(), with some notable differences:
- The names of the returned dataframe are specific to their content. For instance, the column containing the statistic is named following the statistic name, i.e., t, z, etc., instead of a generic name such as statistic.
- It is able to compute or extract indices not available by default, such as p values, CIs, etc.
- It includes feature engineering capabilities, including bootstrapping and standardization of parameters.
Examples
ANOVAs
df <- iris
df$Sepal.Big <- ifelse(df$Sepal.Width >= 3, "Yes", "No")
model <- aov(Sepal.Length ~ Sepal.Big, data = df)
model_parameters(model, eta_squared = "partial")## Parameter | Sum_Squares | df | Mean_Square | F | p | Eta2
## -----------------------------------------------------------------
## Sepal.Big | 1.10 | 1 | 1.10 | 1.61 | 0.207 | 0.01
## Residuals | 101.07 | 148 | 0.68 | | |Mixed models
library(lme4)
model <- lmer(Sepal.Width ~ Petal.Length + Petal.Width + (1|Species), data = iris)
model_parameters(model, standardize = "refit")## Parameter | Coefficient | SE | 95% CI | t(145) | p
## ---------------------------------------------------------------------
## (Intercept) | 3.53e-13 | 1.51 | [-2.96, 2.96] | 2.33e-13 | > .999
## Petal.Length | 0.59 | 0.26 | [ 0.08, 1.10] | 2.26 | 0.024
## Petal.Width | 1.07 | 0.24 | [ 0.60, 1.53] | 4.49 | < .001And you can also directly plot the parameters with the see package!
library(see)
lmer(Sepal.Width ~ Petal.Length * Petal.Width + (1|Species), data = iris) %>%
model_parameters() %>%
plot()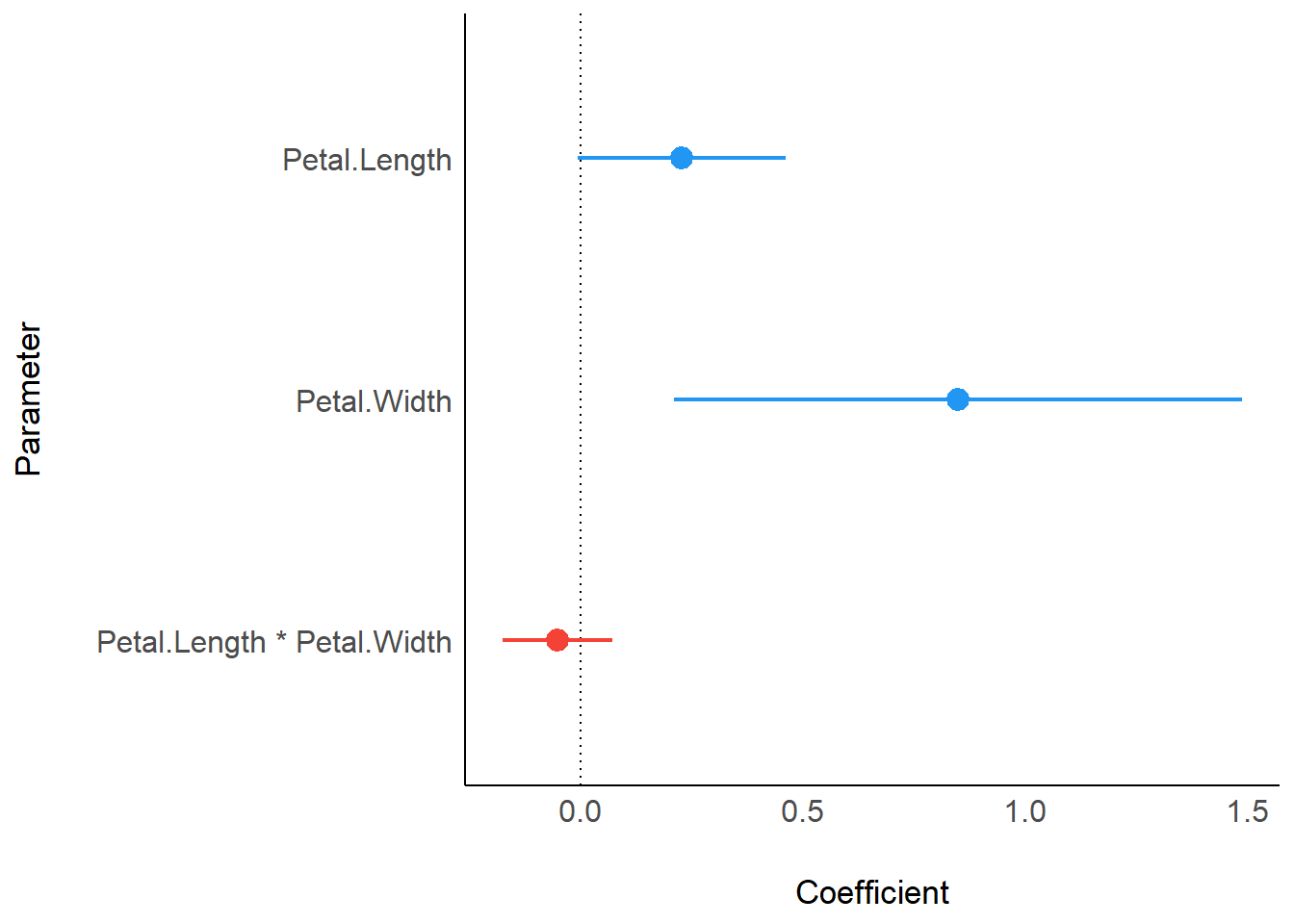
Bayesian models
library(rstanarm)
model <- stan_glm(mpg ~ wt + cyl, data = mtcars)
model_parameters(model)## Parameter | Median | 89% CI | pd | % in ROPE | Rhat | ESS | Prior
## ---------------------------------------------------------------------------------------------------
## (Intercept) | 39.67 | [36.93, 42.68] | 100% | 0% | 0.999 | 4960 | Normal (20.09 +- 15.07)
## wt | -3.19 | [-4.44, -1.92] | 99.98% | 0.10% | 1.002 | 2033 | Normal (0.00 +- 15.40)
## cyl | -1.51 | [-2.18, -0.84] | 99.98% | 1.77% | 1.002 | 2272 | Normal (0.00 +- 8.44)- Check out more examples and documentation here!
Get Involved
There is definitely room for improvement, and some new exciting features are already planned. Feel free to let us know how we could further improve this package!
Note that easystats is a new project in active development, looking for contributors and supporters. Thus, do not hesitate to contact us if you want to get involved :)
- Check out our other blog posts here!
